Age Modelling: Adenoids
Propeller analysis
Gunjan Dixit
February 04, 2025
Last updated: 2025-02-04
Checks: 6 1
Knit directory: paed-airway-allTissues/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of
the R Markdown file created these results, you’ll want to first commit
it to the Git repo. If you’re still working on the analysis, you can
ignore this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230811) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 54e4ec2. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: data/.DS_Store
Ignored: data/RDS/
Ignored: output/.DS_Store
Ignored: output/CSV/.DS_Store
Ignored: output/G000231_Neeland_batch1/
Ignored: output/G000231_Neeland_batch2_1/
Ignored: output/G000231_Neeland_batch2_2/
Ignored: output/G000231_Neeland_batch3/
Ignored: output/G000231_Neeland_batch4/
Ignored: output/G000231_Neeland_batch5/
Ignored: output/G000231_Neeland_batch9_1/
Ignored: output/RDS/
Ignored: output/plots/
Untracked files:
Untracked: analysis/03_Batch_Integration.Rmd
Untracked: analysis/Age_proportions.Rmd
Untracked: analysis/Age_proportions_AllBatches.Rmd
Untracked: analysis/All_Batches_QCExploratory_v2.Rmd
Untracked: analysis/All_metadata.Rmd
Untracked: analysis/Annotation_BAL.Rmd
Untracked: analysis/Annotation_Bronchial_brushings.Rmd
Untracked: analysis/Annotation_Nasal_brushings.Rmd
Untracked: analysis/BatchCorrection_Adenoids.Rmd
Untracked: analysis/BatchCorrection_Nasal_brushings.Rmd
Untracked: analysis/BatchCorrection_Tonsils.Rmd
Untracked: analysis/Batch_Integration_&_Downstream_analysis.Rmd
Untracked: analysis/Batch_correction_&_Downstream.Rmd
Untracked: analysis/Cell_cycle_regression.Rmd
Untracked: analysis/Clustering_Tonsils_v2.Rmd
Untracked: analysis/DGE_analysis_George.Rmd
Untracked: analysis/Master_metadata.Rmd
Untracked: analysis/Pediatric_Vs_Adult_Atlases.Rmd
Untracked: analysis/Preprocessing_Batch1_Nasal_brushings.Rmd
Untracked: analysis/Preprocessing_Batch2_Tonsils.Rmd
Untracked: analysis/Preprocessing_Batch3_Adenoids.Rmd
Untracked: analysis/Preprocessing_Batch4_Bronchial_brushings.Rmd
Untracked: analysis/Preprocessing_Batch5_Nasal_brushings.Rmd
Untracked: analysis/Preprocessing_Batch6_BAL.Rmd
Untracked: analysis/Preprocessing_Batch7_Bronchial_brushings.Rmd
Untracked: analysis/Preprocessing_Batch8_Adenoids.Rmd
Untracked: analysis/Preprocessing_Batch9_Tonsils.Rmd
Untracked: analysis/TonsilsVsAdenoids.Rmd
Untracked: analysis/cell_cycle_regression.R
Untracked: analysis/testing_age_all.Rmd
Untracked: data/Cell_labels_Gunjan_v2/
Untracked: data/Cell_labels_Mel/
Untracked: data/Cell_labels_Mel_v2/
Untracked: data/Cell_labels_Mel_v3/
Untracked: data/Cell_labels_modified_Gunjan/
Untracked: data/Gene_sets/
Untracked: data/Hs.c2.cp.reactome.v7.1.entrez.rds
Untracked: data/Raw_feature_bc_matrix/
Untracked: data/cell_labels_Mel_v4_Dec2024/
Untracked: data/celltypes_Mel_GD_v3.xlsx
Untracked: data/celltypes_Mel_GD_v4_no_dups.xlsx
Untracked: data/celltypes_Mel_modified.xlsx
Untracked: data/celltypes_Mel_v2.csv
Untracked: data/celltypes_Mel_v2.xlsx
Untracked: data/celltypes_Mel_v2_MN.xlsx
Untracked: data/celltypes_for_mel_MN.xlsx
Untracked: data/col_palette.xlsx
Untracked: data/earlyAIR_sample_sheets_combined.xlsx
Untracked: data/~$col_palette.xlsx
Untracked: output/CSV/All_tissues.propeller.xlsx
Untracked: output/CSV/Bronchial_brushings/
Untracked: output/CSV/Bronchial_brushings_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/
Untracked: output/CSV/G000231_Neeland_Adenoids.propeller.xlsx
Untracked: output/CSV/G000231_Neeland_Bronchial_brushings.propeller.xlsx
Untracked: output/CSV/G000231_Neeland_Nasal_brushings.propeller.xlsx
Untracked: output/CSV/G000231_Neeland_Tonsils.propeller.xlsx
Untracked: output/CSV/Nasal_brushings/
Untracked: output/CSV_v2/G000231_Neeland_Adenoids.propeller.xlsx
Untracked: output/CSV_v2/G000231_Neeland_Nasal_brushings.propeller.xlsx
Untracked: output/CSV_v2/G000231_Neeland_Tonsils.propeller.xlsx
Untracked: output/DGE/
Untracked: test_col.csv
Untracked: test_col.txt
Untracked: test_col.xlsx
Unstaged changes:
Deleted: 02_QC_exploratoryPlots.Rmd
Deleted: 02_QC_exploratoryPlots.html
Modified: analysis/00_AllBatches_overview.Rmd
Modified: analysis/01_QC_emptyDrops.Rmd
Modified: analysis/02_QC_exploratoryPlots.Rmd
Modified: analysis/Adenoids.Rmd
Modified: analysis/Adenoids_v2.Rmd
Modified: analysis/Age_modeling.Rmd
Modified: analysis/Age_modelling_Adenoids.Rmd
Modified: analysis/Age_modelling_Nasal_Brushings.Rmd
Modified: analysis/Age_modelling_Tonsils.Rmd
Modified: analysis/AllBatches_QCExploratory.Rmd
Modified: analysis/BAL.Rmd
Modified: analysis/BAL_v2.Rmd
Modified: analysis/Bronchial_brushings.Rmd
Modified: analysis/Bronchial_brushings_v2.Rmd
Modified: analysis/Nasal_brushings.Rmd
Modified: analysis/Nasal_brushings_v2.Rmd
Modified: analysis/Subclustering_Adenoids.Rmd
Modified: analysis/Subclustering_BAL.Rmd
Modified: analysis/Subclustering_Bronchial_brushings.Rmd
Modified: analysis/Subclustering_Nasal_brushings.Rmd
Modified: analysis/Subclustering_Tonsils.Rmd
Modified: analysis/Tonsils.Rmd
Modified: analysis/Tonsils_v2.Rmd
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c0.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c1.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c10.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c11.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c12.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c13.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c14.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c15.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c16.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c17.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c2.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c3.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c4.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c5.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c6.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c7.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c8.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c9.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c0.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c1.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c10.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c11.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c12.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c13.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c14.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c15.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c16.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c17.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c2.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c3.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c4.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c5.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c6.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c7.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c8.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c9.csv
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown
(analysis/Age_modelling_Adenoids.Rmd) and HTML
(docs/Age_modelling_Adenoids.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 8f254aa | Gunjan Dixit | 2024-10-09 | Modified age analysis with interaction model |
| html | 8f254aa | Gunjan Dixit | 2024-10-09 | Modified age analysis with interaction model |
| html | 0f07f72 | Gunjan Dixit | 2024-10-07 | Added BAL subclustering without DecontX |
Introduction
This RMarkdown performs age modelling analysis for the earlyAIR tissue- Adenoids
suppressPackageStartupMessages({
library(here)
library(glue)
library(patchwork)
library(Seurat)
library(dplyr)
library(tidyverse)
library(gridExtra)
library(paletteer)
library(viridis)
library(ggforce)
library(RColorBrewer)
library(scran)
library(ggridges)
library(speckle)
library(edgeR)
library(kableExtra)
library(dplyr)
library(limma)
library(knitr)
library(openxlsx)
})Load data
tissue <- "Adenoids"
out <- here("output/RDS/AllBatches_Annotation_SEUs_v2/G000231_Neeland_Adenoids.annotated_clusters.SEU.rds")
merged_obj <- readRDS(out)
merged_objAn object of class Seurat
17456 features across 184005 samples within 1 assay
Active assay: RNA (17456 features, 2000 variable features)
10 layers present: data.1, data.2, data.3, counts.1, scale.data.1, counts.2, scale.data.2, counts.3, scale.data.3, scale.data
2 dimensional reductions calculated: pca, umap.mergedPlot sample wise cell-type proportions
metadata_df <- data.frame(
sample = merged_obj$sample_id,
donor = merged_obj$donor_id,
age_years = as.character(merged_obj$age_years),
cell_type = Idents(merged_obj)
)
color_palette = readRDS(here("output/RDS/color_palette_unique.rds"))
metadata_df$age_years <- as.numeric(metadata_df$age_years)
barplot_data <- metadata_df %>%
group_by(donor, age_years, cell_type) %>%
summarise(n_cells = n()) %>%
ungroup() %>%
group_by(donor, age_years) %>%
mutate(n_cells_total = sum(n_cells)) %>%
ungroup() %>%
mutate(percentage_cells = n_cells / n_cells_total)`summarise()` has grouped output by 'donor', 'age_years'. You can override
using the `.groups` argument.barplot_data <- barplot_data %>%
arrange(age_years)
a <- ggplot(barplot_data, aes(x = reorder(paste(donor, age_years, sep = ":"), age_years),
y = percentage_cells, fill = cell_type)) +
geom_bar(stat = "identity") +
ggtitle(paste0("Age vs Cell Type Proportions: ", tissue)) +
labs(x = "Sample:Age (Years)", y = "Proportion", fill = "Cell Type") +
scale_fill_manual(values = color_palette) +
theme_minimal() +
theme(
plot.title = element_text(size = 13, hjust = 0.5, face = "bold"),
legend.position = "top",
axis.text.x = element_text(angle = 45, hjust = 1)
)
a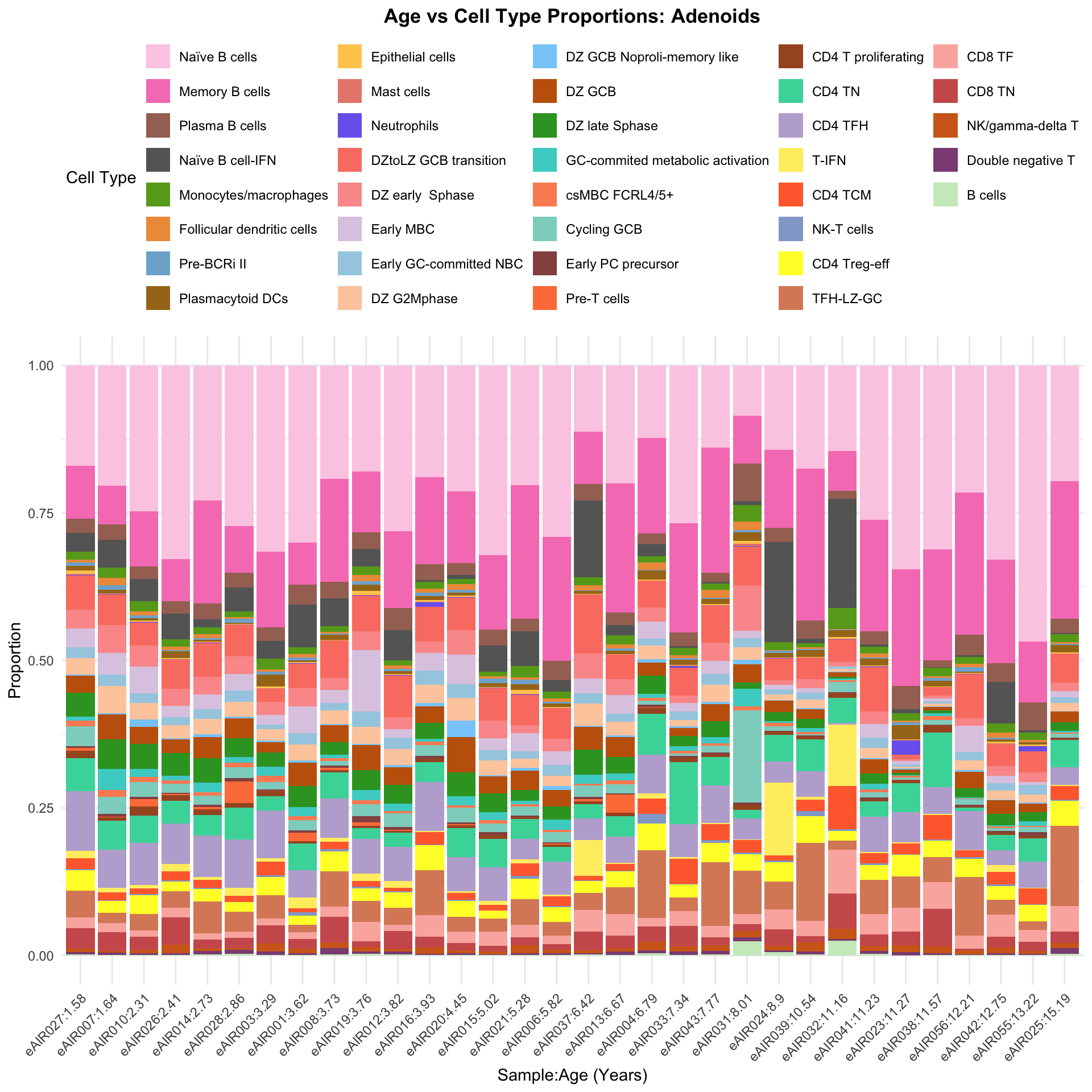
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
Calculate proportions using Propeller
props <- getTransformedProps(clusters = merged_obj$cell_labels_v2,
sample = merged_obj$sample_id, transform = "asin")Performing arcsin square root transformation of proportionscat('### ', tissue, '\n')### Adenoids # Plot Cell Type Mean Variance
p1 <- plotCellTypeMeanVar(props$Counts)Using classic mode.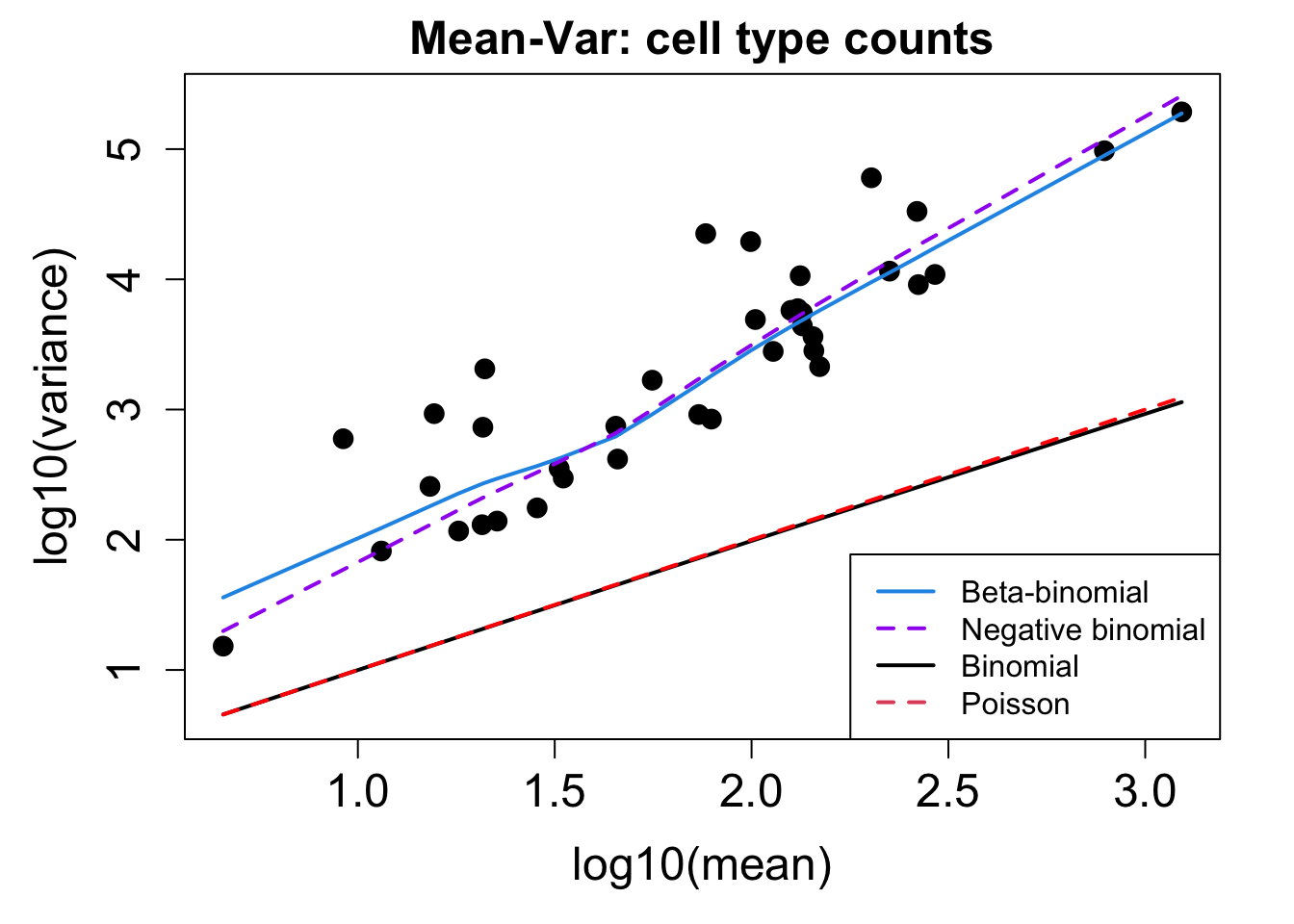
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
# Plot Cell Type Proportions Mean Variance
p2 <- plotCellTypePropsMeanVar(props$Counts)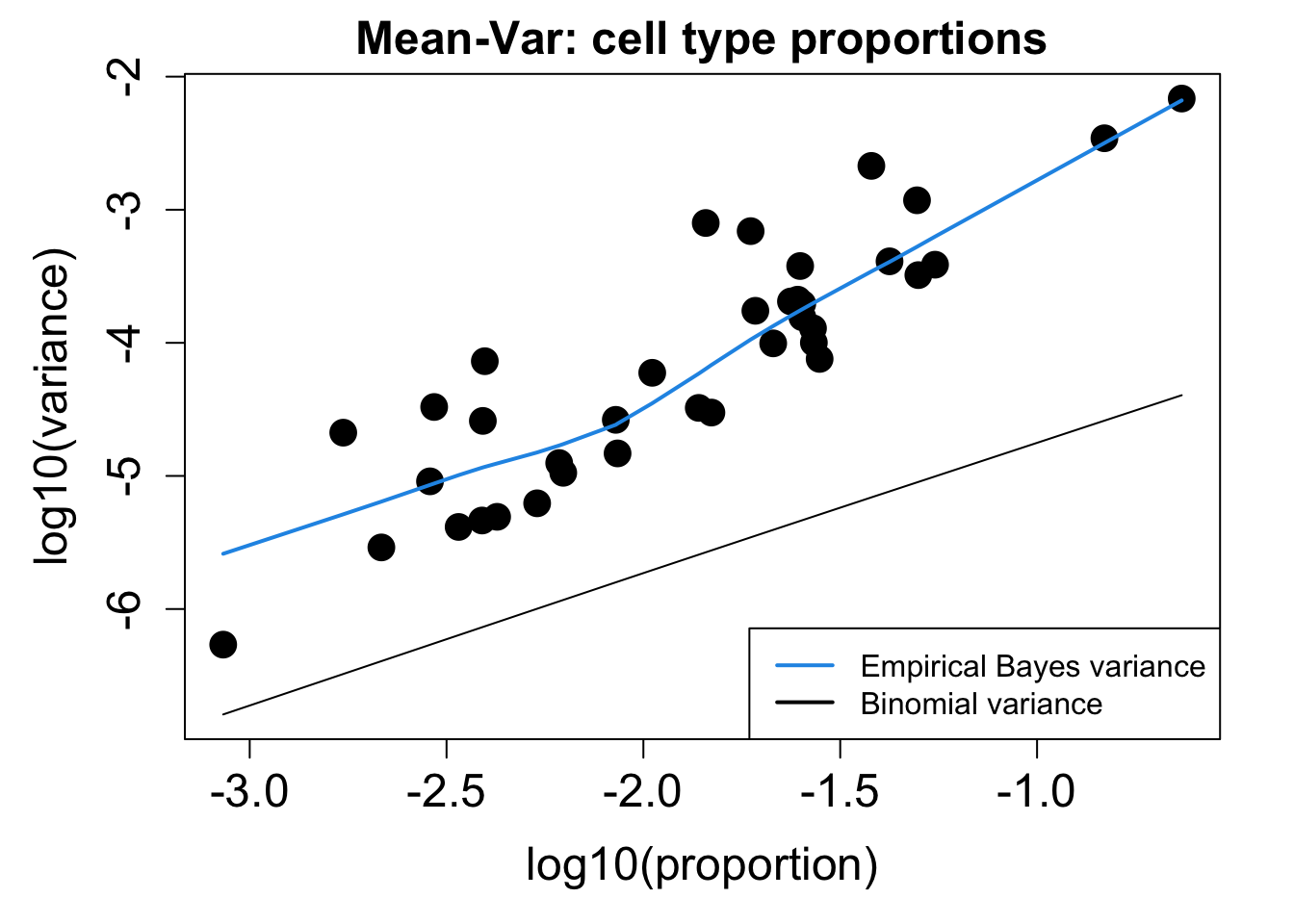
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
p1 / p2numeric(0)print(knitr::kable(props$Proportions, caption = "Cell-type proportions in samples"))
Table: Cell-type proportions in samples
| | s042| s043| s044| s045| s046| s047| s048| s049| s050| s051| s052| s053| s054| s055| s056| s057| s122| s123| s124| s125| s126| s127| s128| s129| s130| s131| s132| s133| s134| s135| s136| s137|
|:--------------------------------|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|
|B cells | 0.0012565| 0.0011970| 0.0036579| 0.0011142| 0.0003168| 0.0016077| 0.0007910| 0.0013493| 0.0011455| 0.0021042| 0.0006300| 0.0006954| 0.0030349| 0.0008116| 0.0006959| 0.0001979| 0.0057615| 0.0023669| 0.0005106| 0.0012739| 0.0027930| 0.0234757| 0.0253067| 0.0010433| 0.0013317| 0.0010542| 0.0021198| 0.0022664| 0.0010437| 0.0016762| 0.0010556| 0.0004396|
|CD4 T proliferating | 0.0041885| 0.0083792| 0.0104108| 0.0037140| 0.0060184| 0.0036748| 0.0158197| 0.0026985| 0.0057274| 0.0097317| 0.0066152| 0.0052156| 0.0037353| 0.0045447| 0.0039434| 0.0045527| 0.0059535| 0.0029586| 0.0097005| 0.0117834| 0.0081796| 0.0088034| 0.0076687| 0.0073031| 0.0042373| 0.0124394| 0.0044628| 0.0039033| 0.0037274| 0.0041905| 0.0042226| 0.0017582|
|CD4 TCM | 0.0069110| 0.0229830| 0.0261677| 0.0155989| 0.0136205| 0.0128617| 0.0096895| 0.0057826| 0.0118366| 0.0136770| 0.0094503| 0.0206885| 0.0094549| 0.0120110| 0.0206449| 0.0178147| 0.0092184| 0.0236686| 0.0154867| 0.0187898| 0.0101746| 0.0208673| 0.0734663| 0.0425143| 0.0077482| 0.0409024| 0.0194131| 0.0177537| 0.0216192| 0.0268194| 0.0269944| 0.0114286|
|CD4 TFH | 0.0462827| 0.0811587| 0.0658413| 0.0560817| 0.0650934| 0.0661461| 0.0711885| 0.0574402| 0.0446735| 0.0699632| 0.0571744| 0.0827538| 0.0582468| 0.0581075| 0.0347947| 0.0506730| 0.0359132| 0.0295858| 0.0689244| 0.1019108| 0.0817955| 0.0352136| 0.0029141| 0.0555556| 0.0364407| 0.0453300| 0.0440701| 0.0600604| 0.0247503| 0.0635564| 0.0438848| 0.0665201|
|CD4 TN | 0.0452356| 0.0237012| 0.0697805| 0.0250696| 0.0489389| 0.0445567| 0.0464702| 0.0246723| 0.0347461| 0.0352446| 0.0480391| 0.0335535| 0.0189098| 0.0495050| 0.0331710| 0.0496833| 0.0453236| 0.0463511| 0.0382914| 0.0560510| 0.0539651| 0.0143463| 0.0426380| 0.1051122| 0.0244552| 0.0923466| 0.0539998| 0.0256862| 0.0262412| 0.0486101| 0.0396622| 0.0058608|
|CD4 Treg-eff | 0.0150785| 0.0304046| 0.0450197| 0.0256267| 0.0199557| 0.0337621| 0.0322325| 0.0264071| 0.0271096| 0.0210416| 0.0133879| 0.0429416| 0.0217112| 0.0271060| 0.0350267| 0.0364212| 0.0326484| 0.0422091| 0.0163376| 0.0350318| 0.0167581| 0.0286925| 0.0165644| 0.0219092| 0.0208232| 0.0274088| 0.0452973| 0.0248048| 0.0237066| 0.0324068| 0.0278993| 0.0313553|
|CD8 TF | 0.0119372| 0.0114915| 0.0140687| 0.0144847| 0.0155211| 0.0176849| 0.0102828| 0.0100231| 0.0370370| 0.0107838| 0.0239408| 0.0368567| 0.0330337| 0.0193150| 0.0213408| 0.0401821| 0.0334166| 0.0435897| 0.0161675| 0.0178344| 0.0114713| 0.0169547| 0.0742331| 0.0255608| 0.0368039| 0.0455408| 0.0263305| 0.0345001| 0.0380200| 0.0192764| 0.0203589| 0.0225641|
|CD8 TN | 0.0165445| 0.0313622| 0.0264491| 0.0094708| 0.0318340| 0.0436380| 0.0217520| 0.0306476| 0.0215731| 0.0144661| 0.0096078| 0.0175591| 0.0096883| 0.0124980| 0.0132220| 0.0237530| 0.0280392| 0.0185404| 0.0457794| 0.0347134| 0.0172569| 0.0114118| 0.0595092| 0.0341680| 0.0319613| 0.0638836| 0.0103760| 0.0188869| 0.0184881| 0.0131303| 0.0149299| 0.0033700|
|csMBC FCRL4/5+ | 0.0069110| 0.0110127| 0.0053461| 0.0042711| 0.0114032| 0.0043638| 0.0085031| 0.0042406| 0.0064910| 0.0055234| 0.0058277| 0.0057371| 0.0067702| 0.0040578| 0.0034795| 0.0029691| 0.0057615| 0.0017751| 0.0062968| 0.0095541| 0.0077805| 0.0068471| 0.0026074| 0.0046948| 0.0032688| 0.0075901| 0.0035702| 0.0050365| 0.0028329| 0.0034921| 0.0025637| 0.0013187|
|Cycling GCB | 0.0171728| 0.0126885| 0.0070343| 0.0168988| 0.0288248| 0.0082683| 0.0251137| 0.0227448| 0.0160367| 0.0160442| 0.0143330| 0.0182545| 0.0204272| 0.0245090| 0.0150777| 0.0039588| 0.0071058| 0.0067061| 0.0153165| 0.0343949| 0.0179551| 0.1565047| 0.0173313| 0.0039124| 0.0188862| 0.0080118| 0.0022314| 0.0081843| 0.0047711| 0.0124319| 0.0081436| 0.0099634|
|Double negative T | 0.0027225| 0.0055063| 0.0045020| 0.0027855| 0.0041178| 0.0103353| 0.0031639| 0.0050116| 0.0055365| 0.0052604| 0.0015751| 0.0020862| 0.0038520| 0.0030839| 0.0025516| 0.0049485| 0.0026887| 0.0098619| 0.0032335| 0.0044586| 0.0066833| 0.0074992| 0.0027607| 0.0057381| 0.0021792| 0.0029517| 0.0045744| 0.0052883| 0.0025347| 0.0061461| 0.0015081| 0.0008791|
|DZ early Sphase | 0.0307853| 0.0217860| 0.0239167| 0.0209842| 0.0476718| 0.0199816| 0.0353965| 0.0210100| 0.0269187| 0.0294582| 0.0300835| 0.0194715| 0.0317497| 0.0415517| 0.0141498| 0.0027712| 0.0082581| 0.0142012| 0.0294418| 0.0324841| 0.0296259| 0.0769482| 0.0073620| 0.0130412| 0.0426150| 0.0050601| 0.0153966| 0.0211534| 0.0165499| 0.0308702| 0.0158347| 0.0124542|
|DZ G2Mphase | 0.0272251| 0.0189131| 0.0230726| 0.0172702| 0.0465632| 0.0231971| 0.0276844| 0.0269854| 0.0236732| 0.0265650| 0.0250433| 0.0309458| 0.0284814| 0.0383055| 0.0157736| 0.0023753| 0.0105627| 0.0138067| 0.0214432| 0.0280255| 0.0251372| 0.0205412| 0.0041411| 0.0106938| 0.0376513| 0.0046384| 0.0146156| 0.0176278| 0.0150589| 0.0284956| 0.0131202| 0.0161172|
|DZ GCB | 0.0387435| 0.0179555| 0.0222285| 0.0276695| 0.0422870| 0.0293983| 0.0294641| 0.0304549| 0.0343643| 0.0355076| 0.0291384| 0.0279903| 0.0424886| 0.0592436| 0.0336349| 0.0065321| 0.0190129| 0.0179487| 0.0234854| 0.0292994| 0.0334165| 0.0313009| 0.0085890| 0.0135629| 0.0386199| 0.0061143| 0.0160660| 0.0239234| 0.0229611| 0.0287750| 0.0147791| 0.0279853|
|DZ GCB Noproli-memory like | 0.0039791| 0.0028729| 0.0056275| 0.0070566| 0.0014254| 0.0018374| 0.0120625| 0.0034695| 0.0024819| 0.0044713| 0.0020476| 0.0060848| 0.0024513| 0.0284045| 0.0044073| 0.0015835| 0.0007682| 0.0003945| 0.0018720| 0.0015924| 0.0029925| 0.0078252| 0.0009202| 0.0023474| 0.0018160| 0.0010542| 0.0016735| 0.0017628| 0.0014910| 0.0037715| 0.0015081| 0.0030769|
|DZ late Sphase | 0.0360209| 0.0241800| 0.0306697| 0.0219127| 0.0503643| 0.0213597| 0.0419221| 0.0319969| 0.0274914| 0.0420831| 0.0315010| 0.0265994| 0.0330337| 0.0409025| 0.0178613| 0.0033650| 0.0149798| 0.0147929| 0.0386317| 0.0401274| 0.0324190| 0.0104336| 0.0029141| 0.0179969| 0.0422518| 0.0046384| 0.0159545| 0.0175019| 0.0187863| 0.0270988| 0.0158347| 0.0153846|
|DZtoLZ GCB transition | 0.0420942| 0.0217860| 0.0453011| 0.0521820| 0.0521064| 0.0627010| 0.0383627| 0.0695837| 0.0412371| 0.0570752| 0.0548118| 0.0584145| 0.0595308| 0.0545366| 0.0510322| 0.0077197| 0.0366814| 0.0487179| 0.0490129| 0.0579618| 0.0526683| 0.0661885| 0.0377301| 0.0469484| 0.0986683| 0.0143369| 0.0355908| 0.0756736| 0.0366781| 0.0628579| 0.0357412| 0.0742857|
|Early GC-committed NBC | 0.0188482| 0.0081398| 0.0112549| 0.0183844| 0.0193221| 0.0117134| 0.0166106| 0.0194680| 0.0129821| 0.0163072| 0.0166955| 0.0241655| 0.0261468| 0.0228859| 0.0118302| 0.0043547| 0.0080661| 0.0104536| 0.0134445| 0.0181529| 0.0193516| 0.0159765| 0.0036810| 0.0146062| 0.0175545| 0.0042167| 0.0120495| 0.0176278| 0.0125242| 0.0181590| 0.0165887| 0.0147985|
|Early MBC | 0.0456545| 0.0162796| 0.0295442| 0.0228412| 0.0364270| 0.0229674| 0.0458770| 0.0138782| 0.0316915| 0.0249868| 0.0203182| 0.0292072| 0.1040037| 0.0506411| 0.0308513| 0.0057403| 0.0078740| 0.0102564| 0.0194010| 0.0318471| 0.0282294| 0.0120639| 0.0004601| 0.0143453| 0.0253027| 0.0046384| 0.0080330| 0.0227902| 0.0126733| 0.0222098| 0.0054290| 0.0441026|
|Early PC precursor | 0.0039791| 0.0023941| 0.0033765| 0.0020427| 0.0047513| 0.0036748| 0.0031639| 0.0048188| 0.0015273| 0.0036823| 0.0045676| 0.0052156| 0.0107389| 0.0030839| 0.0023196| 0.0001979| 0.0007682| 0.0011834| 0.0023826| 0.0028662| 0.0061845| 0.0042387| 0.0015337| 0.0036515| 0.0042373| 0.0006325| 0.0021198| 0.0036515| 0.0029820| 0.0032127| 0.0067863| 0.0026374|
|Epithelial cells | 0.0018848| 0.0026335| 0.0014069| 0.0020427| 0.0006335| 0.0006890| 0.0017797| 0.0021203| 0.0003818| 0.0007891| 0.0011025| 0.0029555| 0.0072371| 0.0025970| 0.0062630| 0.0015835| 0.0009602| 0.0021696| 0.0015317| 0.0060510| 0.0007980| 0.0045647| 0.0029141| 0.0005216| 0.0009685| 0.0010542| 0.0011157| 0.0012591| 0.0025347| 0.0022349| 0.0031669| 0.0011722|
|Follicular dendritic cells | 0.0052356| 0.0038305| 0.0123804| 0.0053853| 0.0120367| 0.0022967| 0.0073166| 0.0057826| 0.0061092| 0.0065755| 0.0039376| 0.0073018| 0.0100385| 0.0094141| 0.0041754| 0.0023753| 0.0003841| 0.0027613| 0.0054459| 0.0047771| 0.0032918| 0.0146723| 0.0016871| 0.0052165| 0.0081114| 0.0025300| 0.0027892| 0.0076807| 0.0087968| 0.0132700| 0.0043734| 0.0055678|
|GC-commited metabolic activation | 0.0157068| 0.0148432| 0.0067530| 0.0176416| 0.0361102| 0.0057418| 0.0142377| 0.0121434| 0.0116457| 0.0160442| 0.0141755| 0.0053894| 0.0173923| 0.0154196| 0.0062630| 0.0013856| 0.0036489| 0.0027613| 0.0073179| 0.0066879| 0.0092768| 0.0293446| 0.0027607| 0.0070423| 0.0153753| 0.0042167| 0.0062479| 0.0094435| 0.0026838| 0.0107557| 0.0069371| 0.0016117|
|Mast cells | 0.0004188| 0.0016758| 0.0000000| 0.0001857| 0.0001584| 0.0004593| 0.0009887| 0.0003855| 0.0005727| 0.0005260| 0.0007875| 0.0010431| 0.0011673| 0.0004869| 0.0009279| 0.0005938| 0.0001920| 0.0005917| 0.0008509| 0.0019108| 0.0009975| 0.0035866| 0.0015337| 0.0010433| 0.0008475| 0.0000000| 0.0004463| 0.0007555| 0.0020874| 0.0004191| 0.0001508| 0.0016117|
|Memory B cells | 0.0718325| 0.1288006| 0.1623523| 0.2102136| 0.0657270| 0.1740928| 0.0943247| 0.1303007| 0.2185949| 0.1743819| 0.1263191| 0.1470793| 0.1028365| 0.1212466| 0.2261656| 0.1983373| 0.1319378| 0.2333333| 0.0713070| 0.0891720| 0.0792020| 0.0808608| 0.0673313| 0.1851852| 0.0880145| 0.1884883| 0.2571684| 0.1884916| 0.1756374| 0.2120408| 0.1034535| 0.2411722|
|Monocytes/macrophages | 0.0115183| 0.0189131| 0.0101294| 0.0120706| 0.0175800| 0.0098760| 0.0174016| 0.0080956| 0.0158457| 0.0115729| 0.0077178| 0.0116481| 0.0148243| 0.0089271| 0.0192531| 0.0124703| 0.0136355| 0.0136095| 0.0124234| 0.0133758| 0.0097756| 0.0283665| 0.0352761| 0.0125196| 0.0142857| 0.0141261| 0.0066942| 0.0107026| 0.0156553| 0.0106160| 0.0119137| 0.0115751|
|Naïve B cell-IFN | 0.0730890| 0.0299258| 0.0205402| 0.0189415| 0.0476718| 0.0473128| 0.0377694| 0.0516577| 0.0173730| 0.0136770| 0.0444164| 0.0027816| 0.0295319| 0.0063301| 0.0584551| 0.0071259| 0.1697715| 0.0025641| 0.0435671| 0.0321656| 0.0403990| 0.0061950| 0.1851227| 0.0028691| 0.1301453| 0.0016867| 0.0080330| 0.0045329| 0.0708215| 0.0026540| 0.0039210| 0.0030769|
|Naïve B cells | 0.2998953| 0.3155375| 0.1226787| 0.2909935| 0.2035160| 0.1926964| 0.2467866| 0.2812259| 0.2002673| 0.2293530| 0.3213104| 0.1898470| 0.1799930| 0.2134394| 0.2032011| 0.3452098| 0.1430766| 0.1964497| 0.3286249| 0.1703822| 0.2726185| 0.0854255| 0.1452454| 0.2670840| 0.1127119| 0.3116171| 0.1750530| 0.2618988| 0.3293574| 0.1395446| 0.4676519| 0.2150916|
|Neutrophils | 0.0004188| 0.0000000| 0.0000000| 0.0001857| 0.0001584| 0.0000000| 0.0007910| 0.0011565| 0.0000000| 0.0015781| 0.0006300| 0.0081711| 0.0008171| 0.0006492| 0.0009279| 0.0243468| 0.0005761| 0.0000000| 0.0003404| 0.0006369| 0.0001995| 0.0009782| 0.0004601| 0.0028691| 0.0001211| 0.0000000| 0.0000000| 0.0001259| 0.0000000| 0.0002794| 0.0085960| 0.0002930|
|NK-T cells | 0.0058639| 0.0035911| 0.0160383| 0.0022284| 0.0006335| 0.0039045| 0.0017797| 0.0015420| 0.0009546| 0.0023672| 0.0001575| 0.0008693| 0.0030349| 0.0017854| 0.0048713| 0.0013856| 0.0028807| 0.0017751| 0.0020422| 0.0015924| 0.0001995| 0.0026084| 0.0021472| 0.0010433| 0.0009685| 0.0023192| 0.0092603| 0.0028960| 0.0022365| 0.0051683| 0.0015081| 0.0021978|
|NK/gamma-delta T | 0.0071204| 0.0129279| 0.0143500| 0.0055710| 0.0033259| 0.0096463| 0.0059324| 0.0046261| 0.0053456| 0.0052604| 0.0044101| 0.0109527| 0.0073538| 0.0045447| 0.0139179| 0.0116785| 0.0076820| 0.0092702| 0.0146358| 0.0060510| 0.0025935| 0.0107597| 0.0173313| 0.0086072| 0.0047215| 0.0109635| 0.0152851| 0.0089398| 0.0093932| 0.0097779| 0.0054290| 0.0067399|
|Plasma B cells | 0.0332984| 0.0225042| 0.0182893| 0.0323120| 0.0259740| 0.0280202| 0.0203678| 0.0368157| 0.0211913| 0.0265650| 0.0274059| 0.0265994| 0.0283647| 0.0207758| 0.0215727| 0.0389945| 0.0243902| 0.0234714| 0.0211028| 0.0238854| 0.0243392| 0.0639061| 0.0134969| 0.0232134| 0.0277240| 0.0109635| 0.0309048| 0.0221607| 0.0310124| 0.0155050| 0.0475041| 0.0347253|
|Plasmacytoid DCs | 0.0027225| 0.0203495| 0.0157569| 0.0051996| 0.0063351| 0.0089573| 0.0041527| 0.0042406| 0.0061092| 0.0031562| 0.0066152| 0.0067803| 0.0107389| 0.0050317| 0.0099745| 0.0241489| 0.0078740| 0.0122288| 0.0115725| 0.0076433| 0.0025935| 0.0143463| 0.0113497| 0.0088680| 0.0049637| 0.0090660| 0.0120495| 0.0114581| 0.0043238| 0.0092192| 0.0031669| 0.0013187|
|Pre-BCRi II | 0.0048168| 0.0043093| 0.0011255| 0.0035283| 0.0077605| 0.0025264| 0.0045482| 0.0044333| 0.0042001| 0.0028932| 0.0070877| 0.0053894| 0.0057196| 0.0048693| 0.0074229| 0.0047506| 0.0036489| 0.0017751| 0.0020422| 0.0057325| 0.0060848| 0.0039126| 0.0009202| 0.0028691| 0.0012107| 0.0054818| 0.0013388| 0.0018887| 0.0020874| 0.0016762| 0.0003016| 0.0082051|
|Pre-T cells | 0.0140314| 0.0004788| 0.0008441| 0.0024141| 0.0006335| 0.0039045| 0.0126557| 0.0019275| 0.0309278| 0.0034193| 0.0000000| 0.0000000| 0.0058363| 0.0021100| 0.0000000| 0.0003959| 0.0000000| 0.0000000| 0.0015317| 0.0047771| 0.0364090| 0.0000000| 0.0001534| 0.0000000| 0.0039952| 0.0000000| 0.0000000| 0.0001259| 0.0000000| 0.0000000| 0.0000000| 0.0000000|
|T-IFN | 0.0178010| 0.0057458| 0.0087226| 0.0027855| 0.0079189| 0.0066605| 0.0057346| 0.0121434| 0.0013364| 0.0044713| 0.0070877| 0.0027816| 0.0126065| 0.0022724| 0.0074229| 0.0023753| 0.1229115| 0.0019724| 0.0124234| 0.0117834| 0.0133666| 0.0013042| 0.1042945| 0.0028691| 0.0602906| 0.0021084| 0.0040165| 0.0012591| 0.0113314| 0.0015365| 0.0009048| 0.0005861|
|TFH-LZ-GC | 0.0127749| 0.0397414| 0.1153630| 0.0228412| 0.0172632| 0.0594855| 0.0278821| 0.0287201| 0.0448645| 0.0533930| 0.0220507| 0.0759736| 0.0350181| 0.0249959| 0.0433774| 0.0530483| 0.0476282| 0.1358974| 0.0280803| 0.0452229| 0.0330175| 0.0730355| 0.0156442| 0.0234742| 0.0286925| 0.0425891| 0.1316523| 0.0582977| 0.0246012| 0.1081157| 0.0141758| 0.0987546|Set model and design matrix
merged_obj@meta.data <- merged_obj@meta.data %>%
mutate(age_group = case_when(
age_years >= 1 & age_years < 6 ~ "Preschool_1to5_years",
age_years >= 6 & age_years < 12 ~ "Kids_6to11_years",
age_years >= 12 ~ "Adolescent_12to17_years",
TRUE ~ "Other"))
samples_metadata <- merged_obj@meta.data %>%
dplyr::filter(sample_id %in% unique(merged_obj@meta.data$sample_id)) %>%
dplyr::group_by(sample_id) %>%
dplyr::summarise(
age = dplyr::first(age_years),
sex = dplyr::first(sex),
batch = dplyr::first(batch_name),
age_group = dplyr::first(age_group),
.groups = 'drop'
)
age <- samples_metadata$age
sex <- as.factor(samples_metadata$sex)
batch <- as.factor(samples_metadata$batch)
design <- model.matrix(~age + sex + batch)
design (Intercept) age sexM batchG000231_batch8
1 1 3.62 1 0
2 1 3.29 0 0
3 1 6.79 0 0
4 1 5.82 0 0
5 1 1.64 1 0
6 1 3.73 1 0
7 1 2.31 1 0
8 1 3.82 1 0
9 1 6.67 0 0
10 1 2.73 1 0
11 1 5.02 0 0
12 1 3.93 1 0
13 1 3.76 1 0
14 1 4.45 1 0
15 1 5.28 0 0
16 1 11.27 0 0
17 1 8.90 1 1
18 1 15.19 1 1
19 1 2.41 1 1
20 1 1.58 0 1
21 1 2.86 0 1
22 1 8.01 1 1
23 1 11.16 1 1
24 1 7.34 0 1
25 1 6.42 1 1
26 1 11.57 1 1
27 1 10.54 0 1
28 1 11.23 1 1
29 1 12.75 1 1
30 1 7.77 0 1
31 1 13.22 0 1
32 1 12.21 0 1
attr(,"assign")
[1] 0 1 2 3
attr(,"contrasts")
attr(,"contrasts")$sex
[1] "contr.treatment"
attr(,"contrasts")$batch
[1] "contr.treatment"Linear model fitting with Limma
fit <- lmFit(props$TransformedProps, design)
fit <- eBayes(fit, robust=TRUE)
coef = "age"
toptable.transformedProps <- topTable(fit, coef = coef)
fit.prop <- lmFit(props$Proportions, design)
fit.prop <- eBayes(fit.prop, robust=TRUE)
toptable.props <- topTable(fit.prop, sort.by = "p", coef = 2)
cat(paste('### ', tissue, '\n', sep = ""))### Adenoidsprint(knitr::kable(toptable.transformedProps, caption = paste0("Transformed proportions Toptable results: ", tissue)))
Table: Transformed proportions Toptable results: Adenoids
| | logFC| AveExpr| t| P.Value| adj.P.Val| B|
|:--------------------------------|----------:|---------:|---------:|---------:|---------:|---------:|
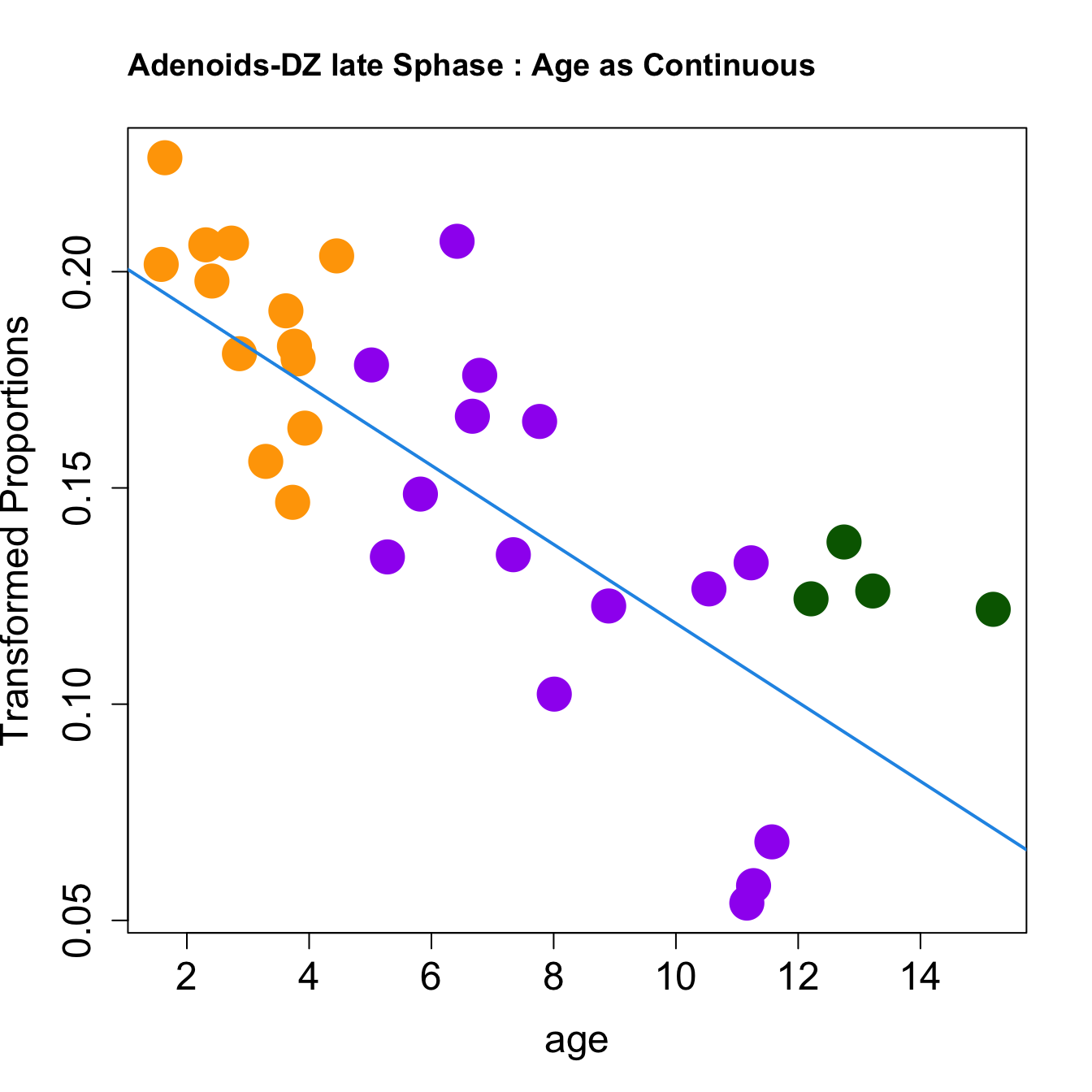
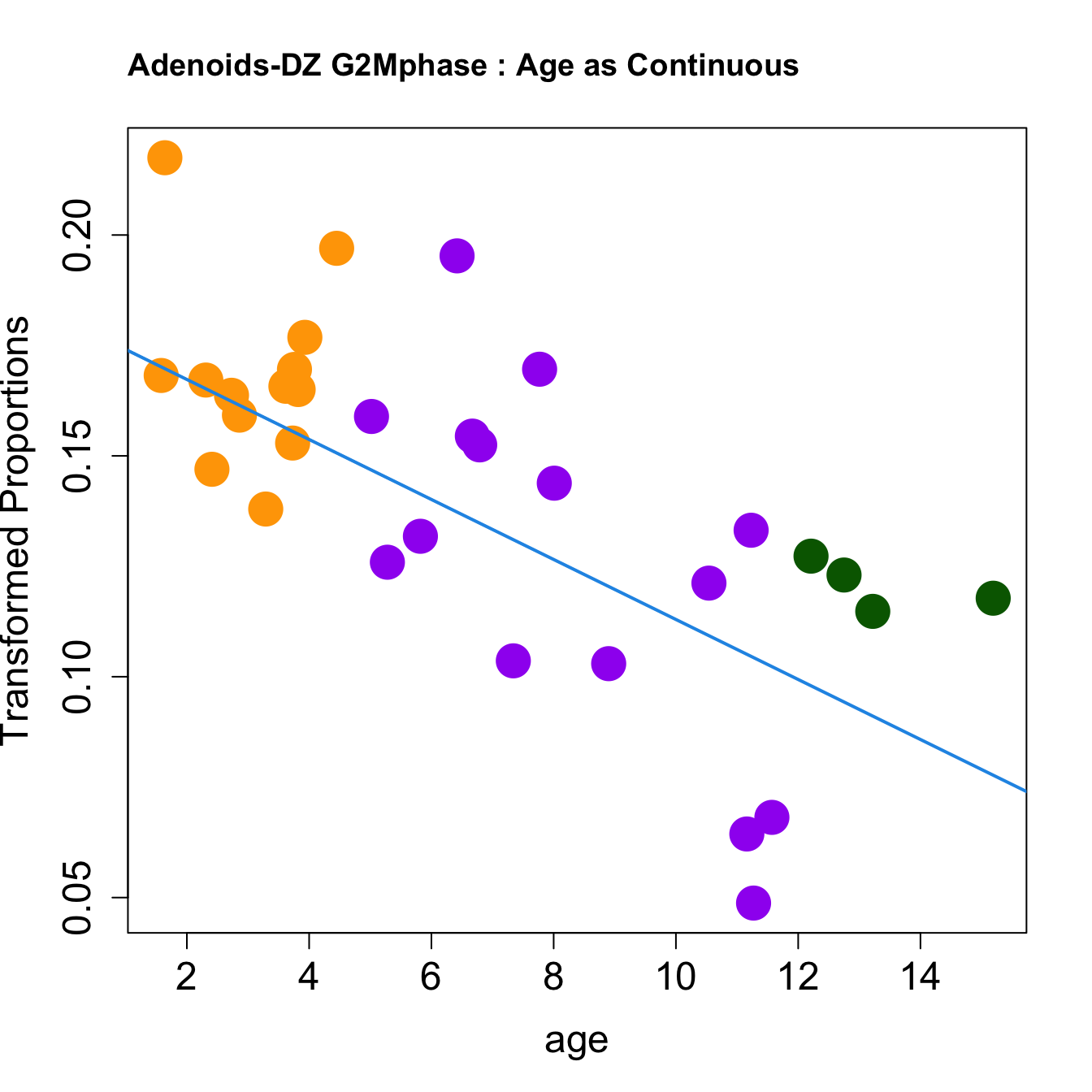
|DZ late Sphase | -0.0091283| 0.1540219| -5.604706| 0.0000039| 0.0001445| 2.372546|
|DZ G2Mphase | -0.0067936| 0.1420384| -4.379118| 0.0001280| 0.0019915| -1.104915|
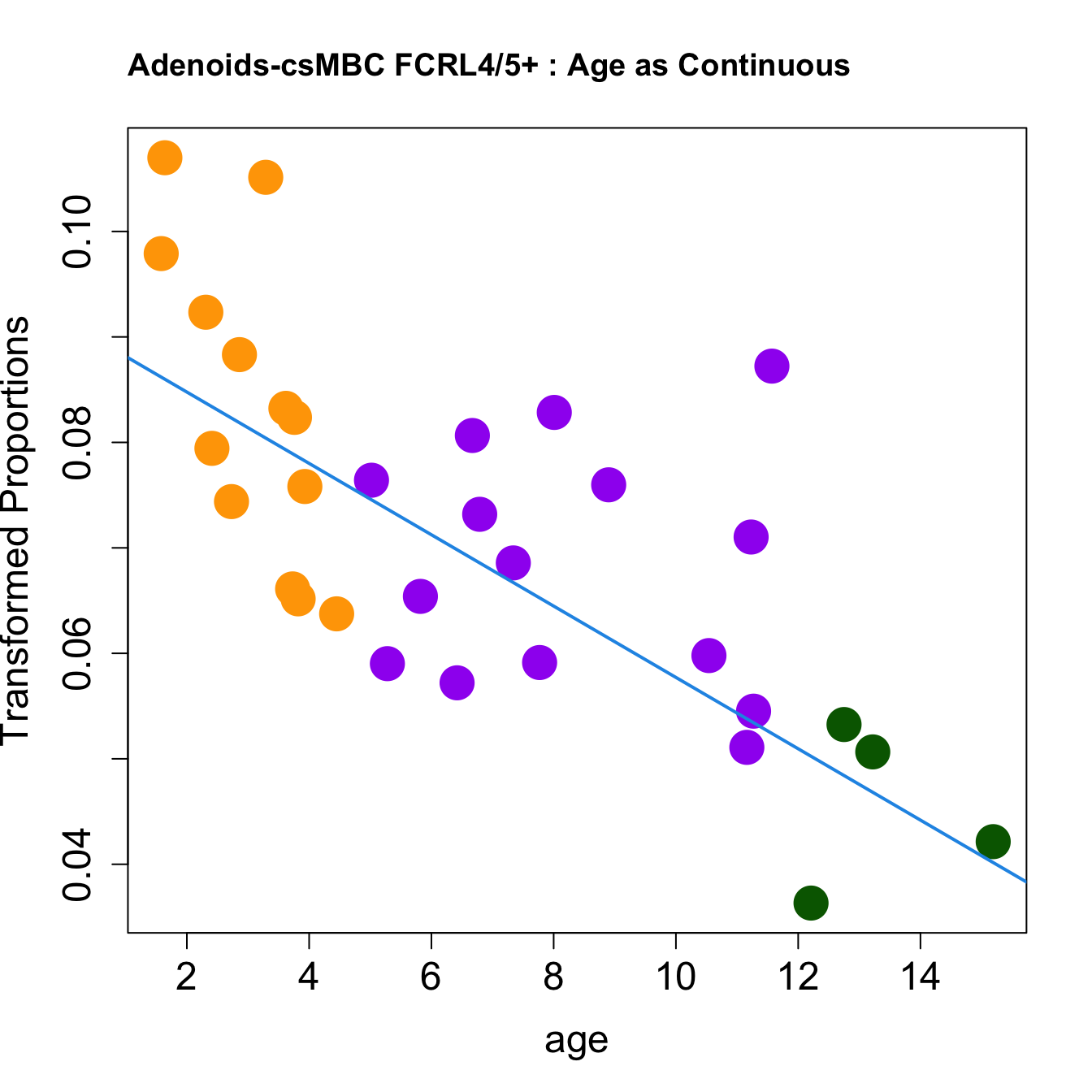
|csMBC FCRL4/5+ | -0.0033821| 0.0714178| -4.202144| 0.0002105| 0.0019915| -1.595011|
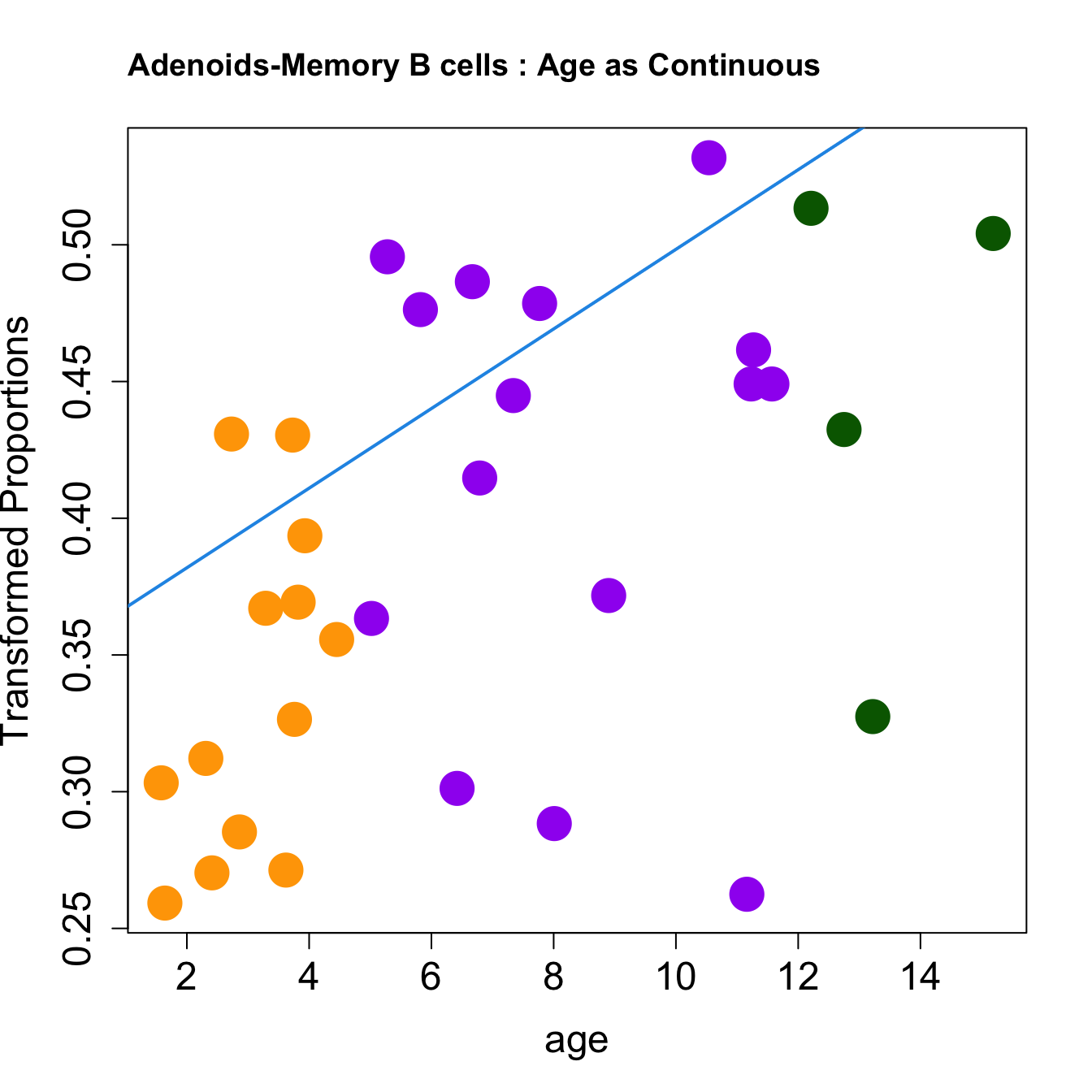
|Memory B cells | 0.0145601| 0.3883601| 4.161395| 0.0002387| 0.0019915| -1.716354|
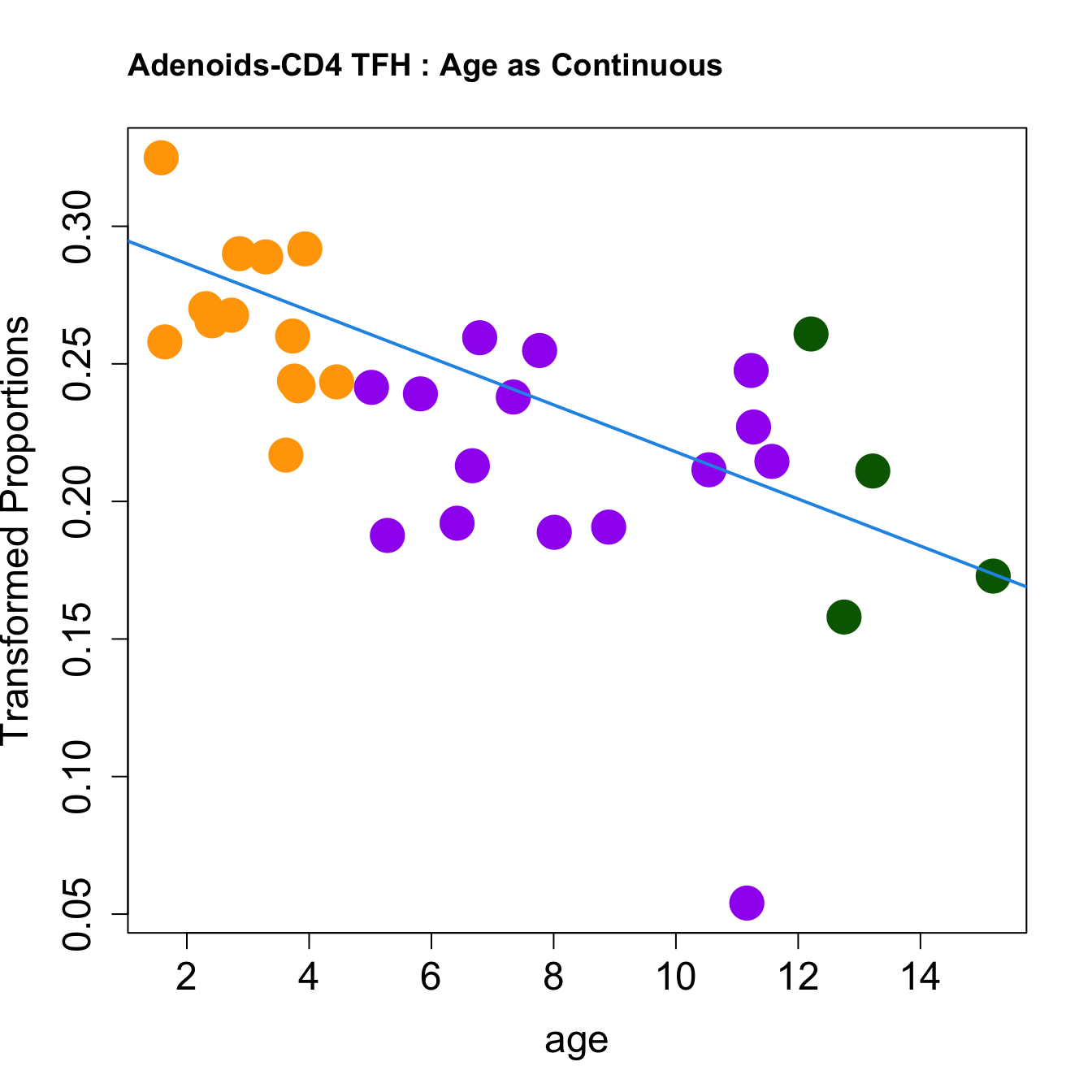
|CD4 TFH | -0.0085537| 0.2320508| -4.114150| 0.0002691| 0.0019915| -1.836690|
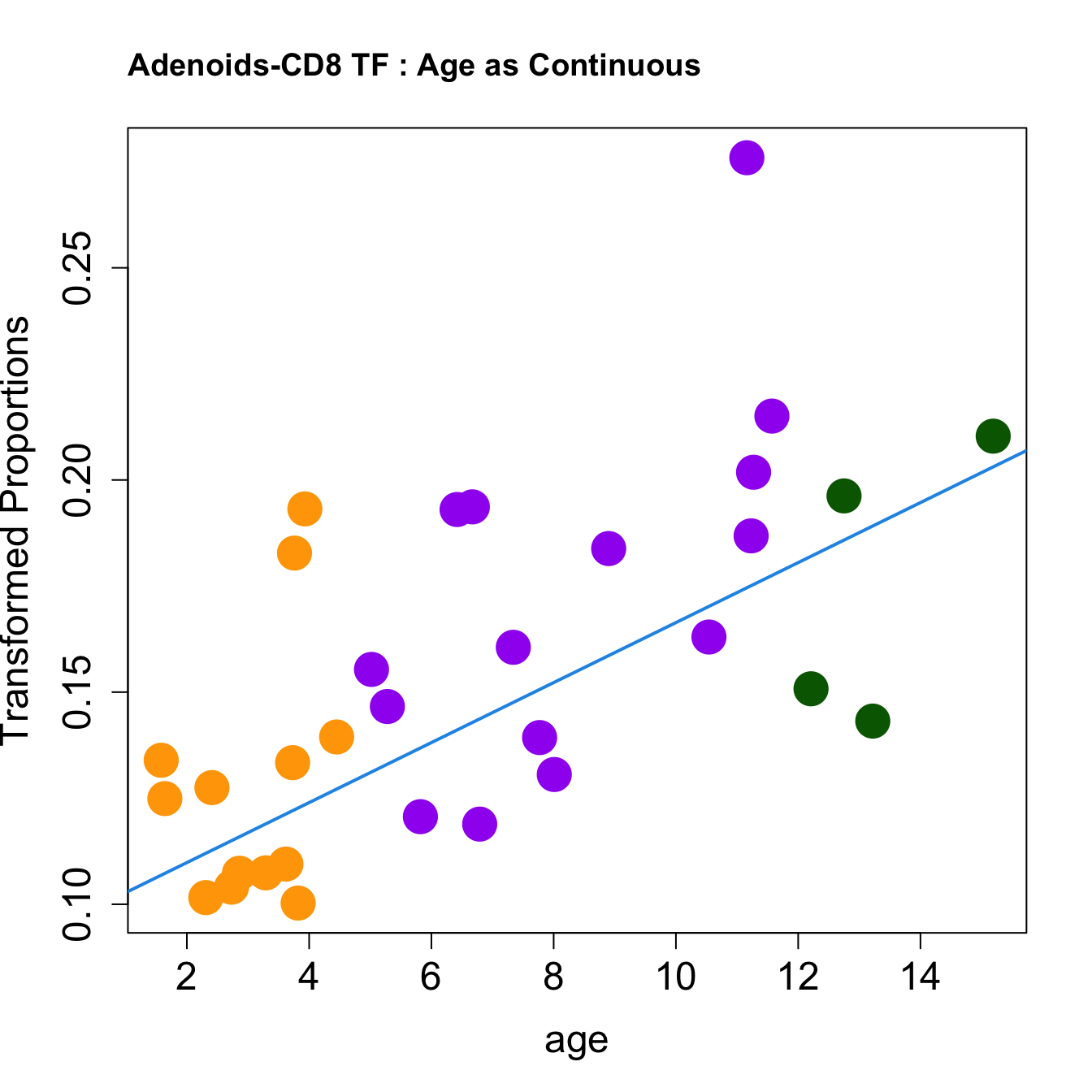
|CD8 TF | 0.0070714| 0.1547135| 4.046547| 0.0003248| 0.0020031| -2.021358|
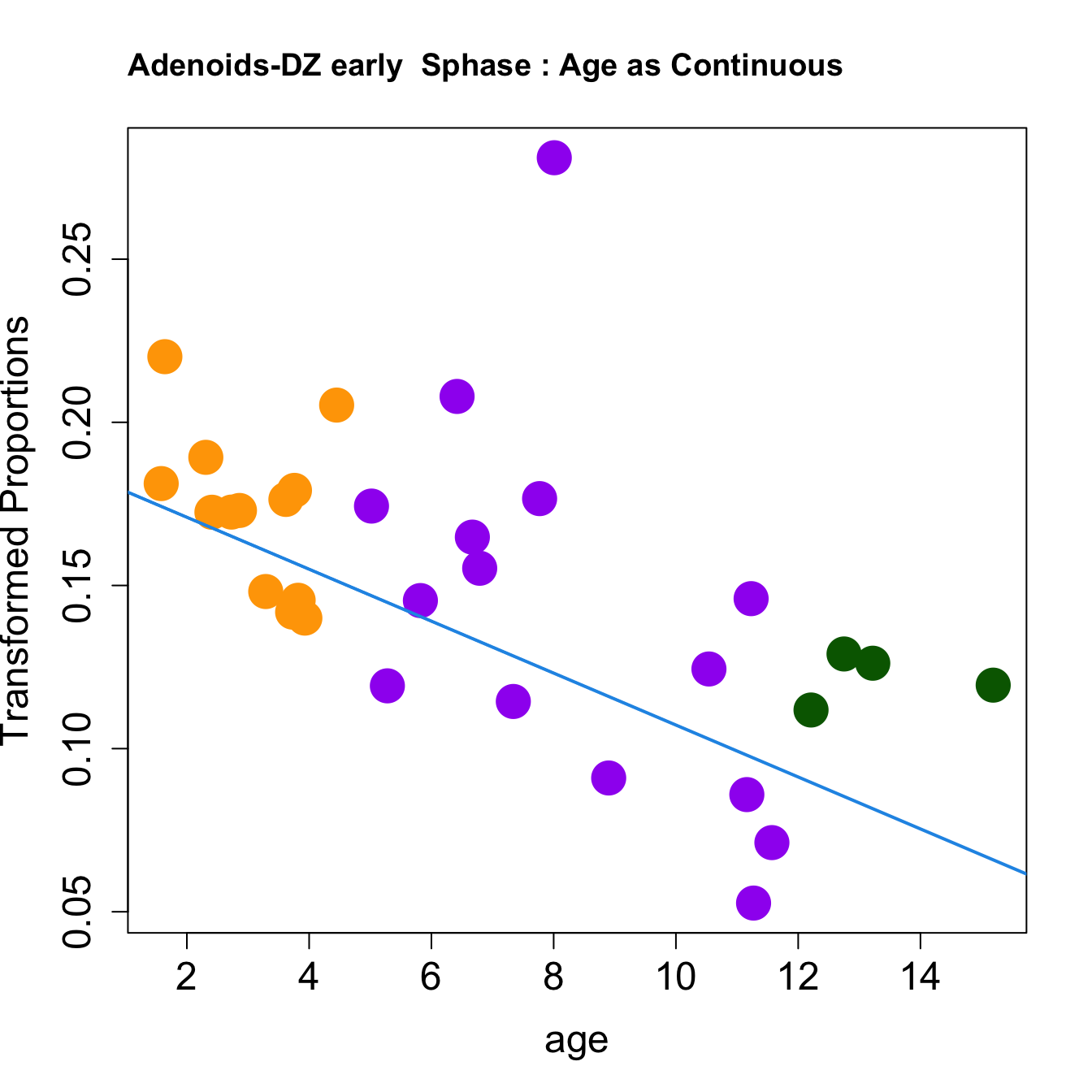
|DZ early Sphase | -0.0079620| 0.1512945| -3.846497| 0.0005645| 0.0029839| -2.562110|
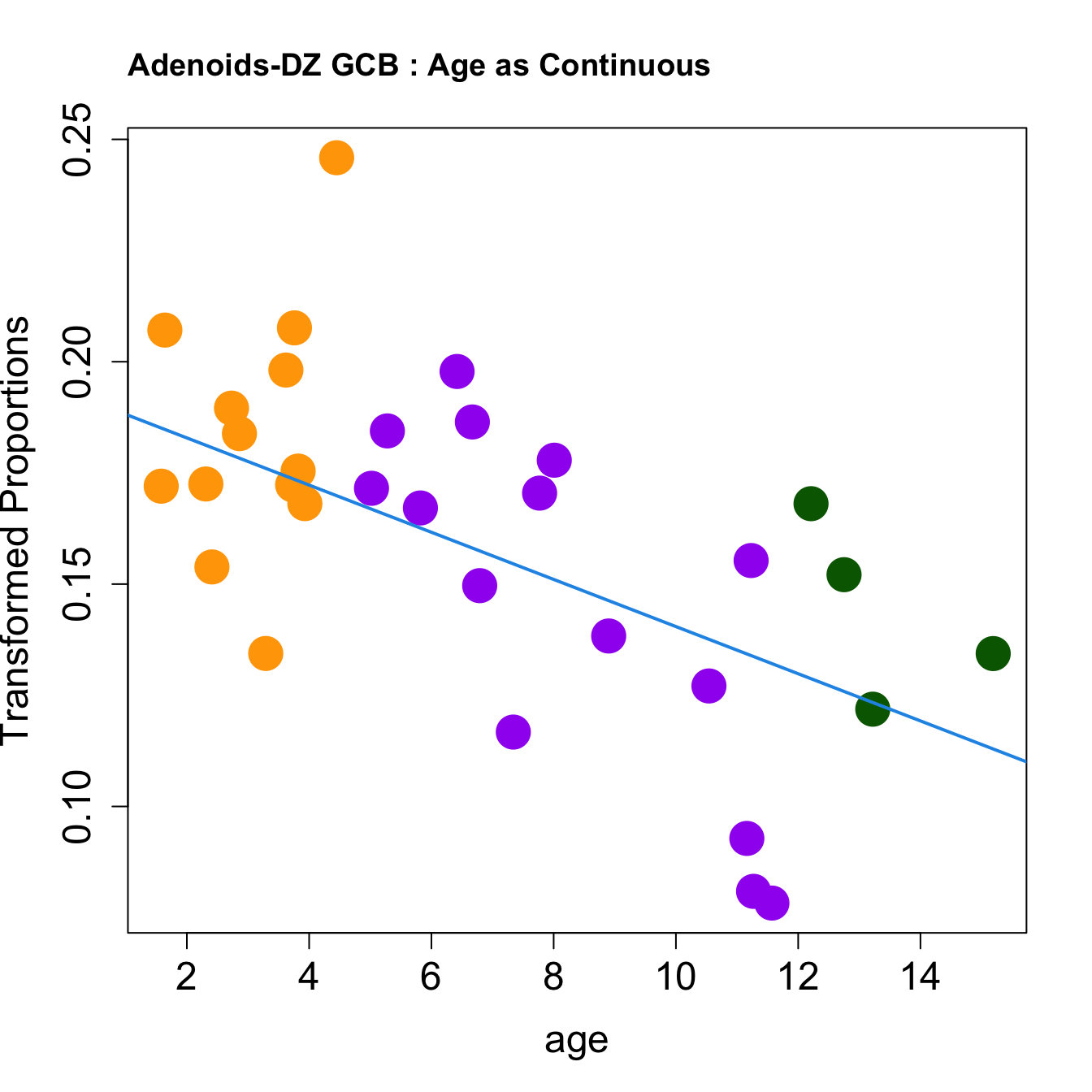
|DZ GCB | -0.0053032| 0.1609978| -3.219026| 0.0030307| 0.0140171| -4.187079|
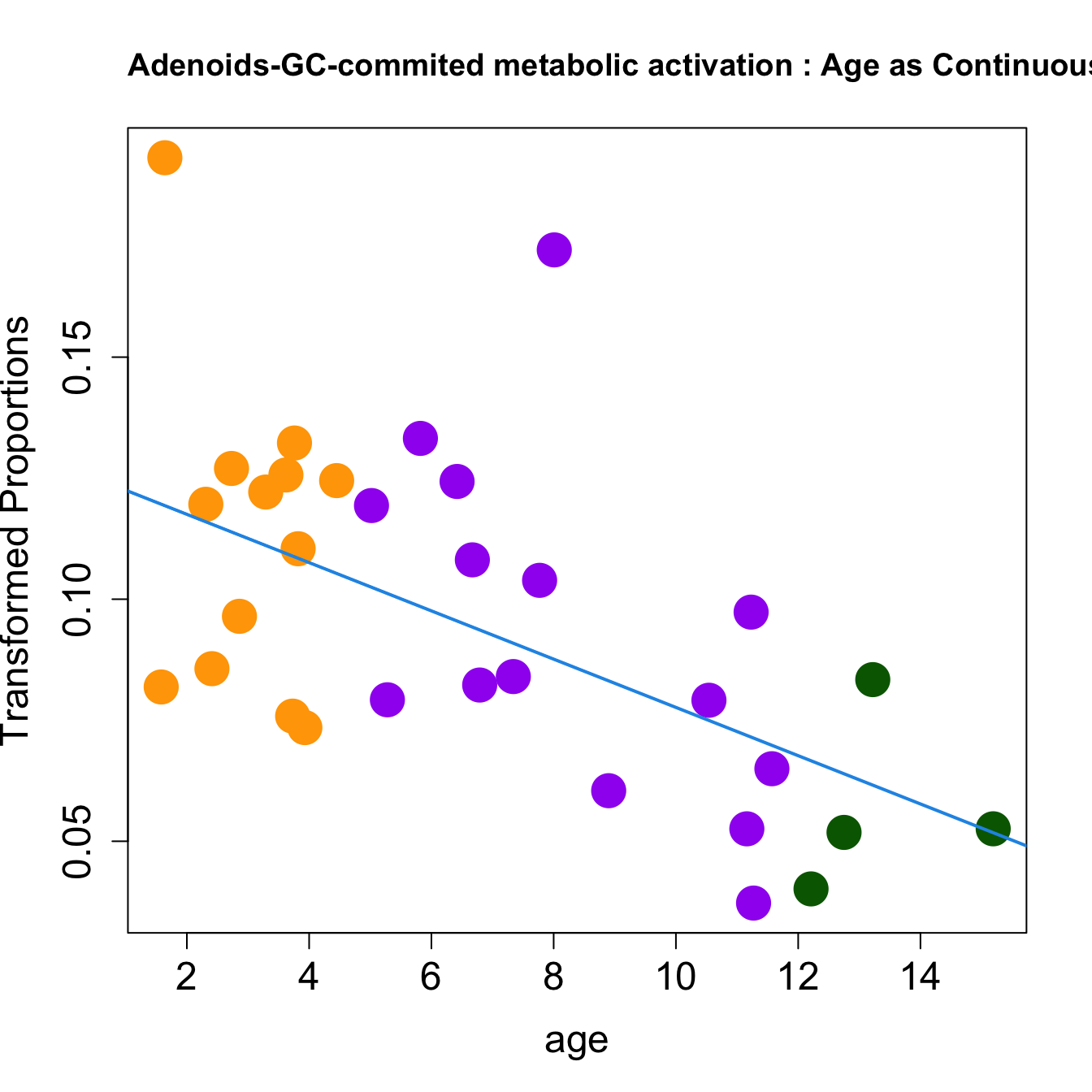
|GC-commited metabolic activation | -0.0049868| 0.0966337| -3.032760| 0.0048917| 0.0201103| -4.643030|
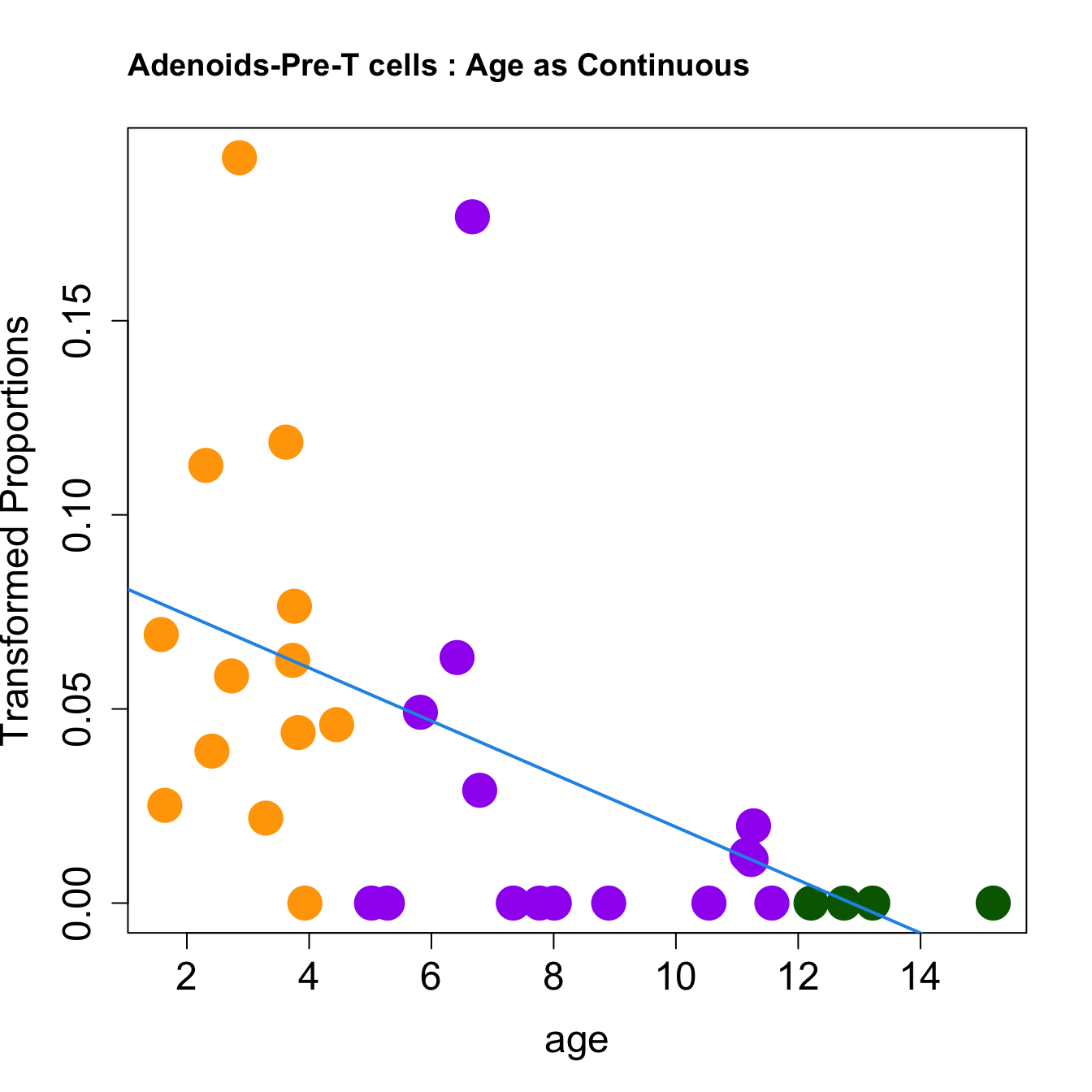
|Pre-T cells | -0.0068313| 0.0383735| -2.758206| 0.0097032| 0.0340046| -5.287862|print(knitr::kable(toptable.props, caption = paste0("Proportions Toptable results: ", tissue)))
Table: Proportions Toptable results: Adenoids
| | logFC| AveExpr| t| P.Value| adj.P.Val| B|
|:----------------------|----------:|---------:|---------:|---------:|---------:|---------:|
|DZ late Sphase | -0.0026081| 0.0253441| -5.957600| 0.0000018| 0.0000663| 1.451431|
|CD4 TFH | -0.0037602| 0.0550639| -4.779690| 0.0000468| 0.0008658| -1.822607|
|DZ G2Mphase | -0.0017237| 0.0213672| -4.293384| 0.0001795| 0.0022138| -3.156687|
|csMBC FCRL4/5+ | -0.0004774| 0.0053718| -4.096114| 0.0003067| 0.0024490| -3.685954|
|Memory B cells | 0.0100725| 0.1482938| 4.069820| 0.0003309| 0.0024490| -3.759882|
|CD8 TF | 0.0022942| 0.0253315| 3.824678| 0.0006427| 0.0039636| -4.410879|
|DZ early Sphase | -0.0022006| 0.0246557| -3.156666| 0.0037064| 0.0195908| -6.106548|
|DZ GCB | -0.0014881| 0.0269669| -2.967503| 0.0059633| 0.0275802| -6.559260|
|Early GC-committed NBC | -0.0007410| 0.0148863| -2.705660| 0.0112844| 0.0463915| -7.159900|
|CD4 T proliferating | -0.0004359| 0.0062582| -2.484113| 0.0189912| 0.0702673| -7.642512|get_age_group_color <- function(age) {
if (age >= 1 && age <= 5) {
return("orange") # Preschool (1-5 years)
} else if (age > 5 && age <= 12) {
return("purple") # Kids (6-11 years)
} else if (age > 12 && age <= 17) {
return("darkgreen") # Adolescent (12-17 years)
} else {
return("black") # Default color for other cases
}
}
age_group_colors <- sapply(age, get_age_group_color)
par(mfrow=c(1,1))
for (i in rownames(toptable.transformedProps)) {
plot(age, props$TransformedProps[i,],
pch=16, cex=3, ylab="Transformed Proportions", cex.lab=1.5, cex.axis=1.5,cex.main=2, col=age_group_colors)
abline(a=fit$coefficients[i, 1], b=fit$coefficients[i, 2], col=4, lwd=2)
title(paste0(tissue, "-", i, " : Age as Continuous"), cex.main = 1.2, adj = 0)
}
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
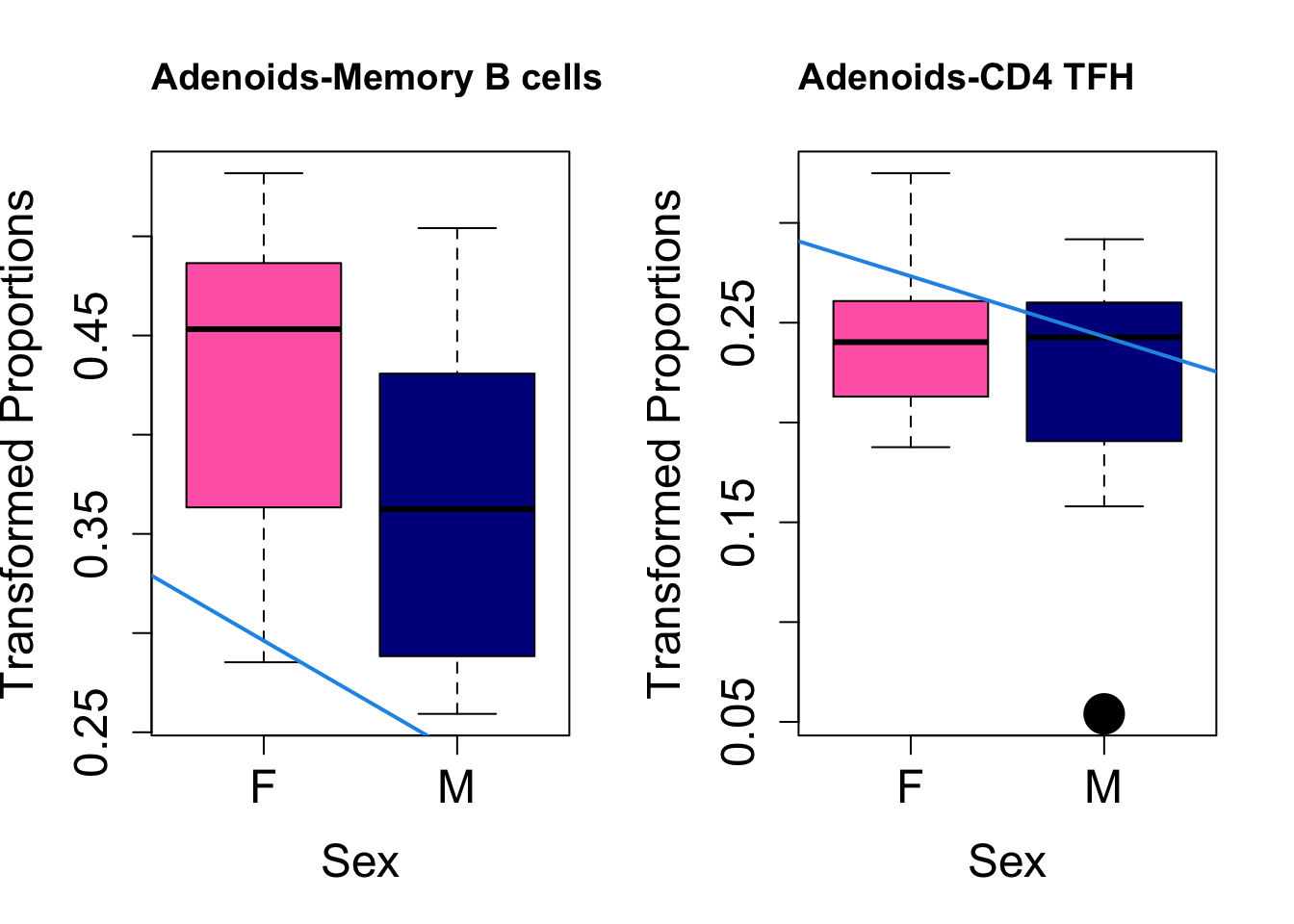
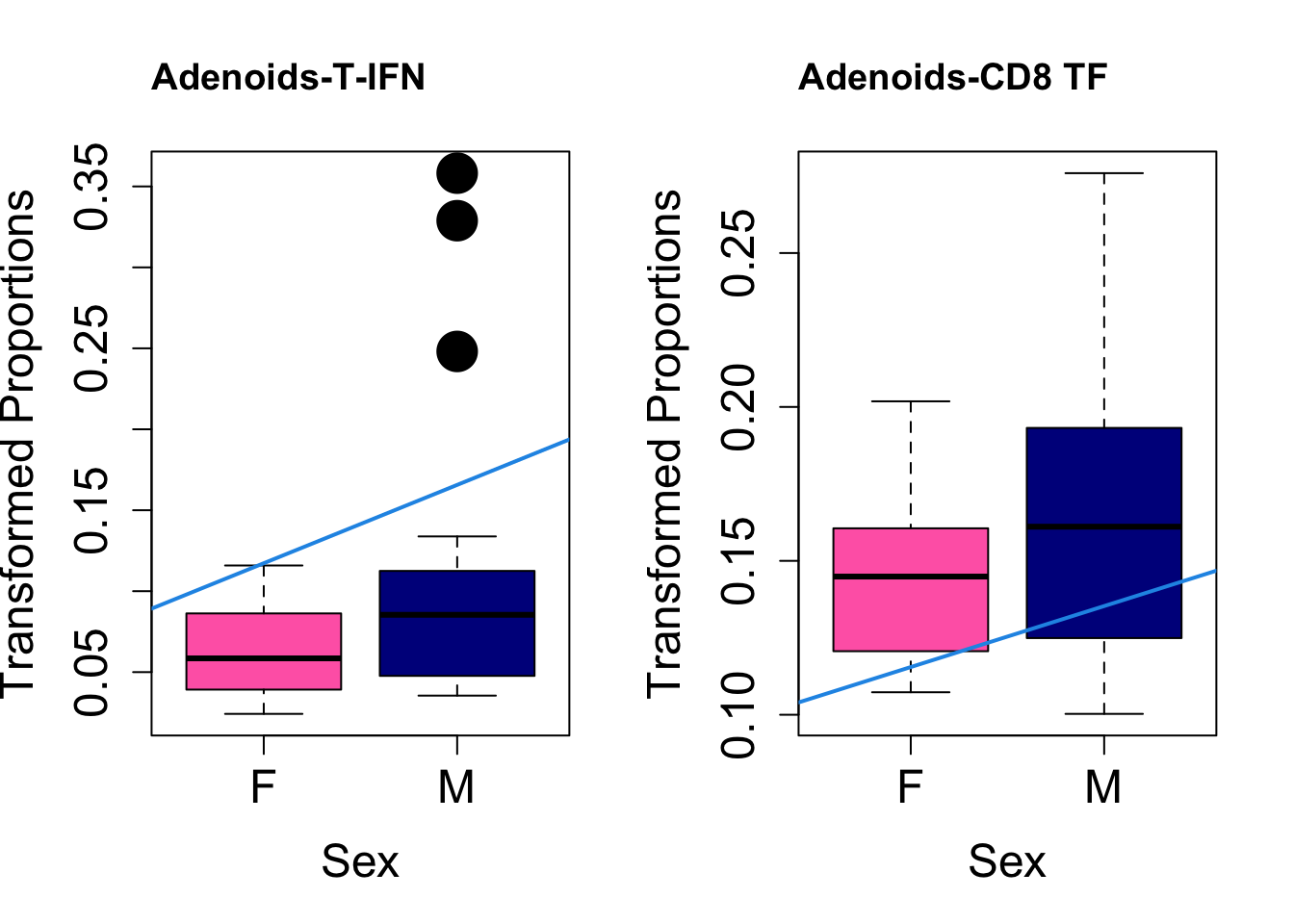
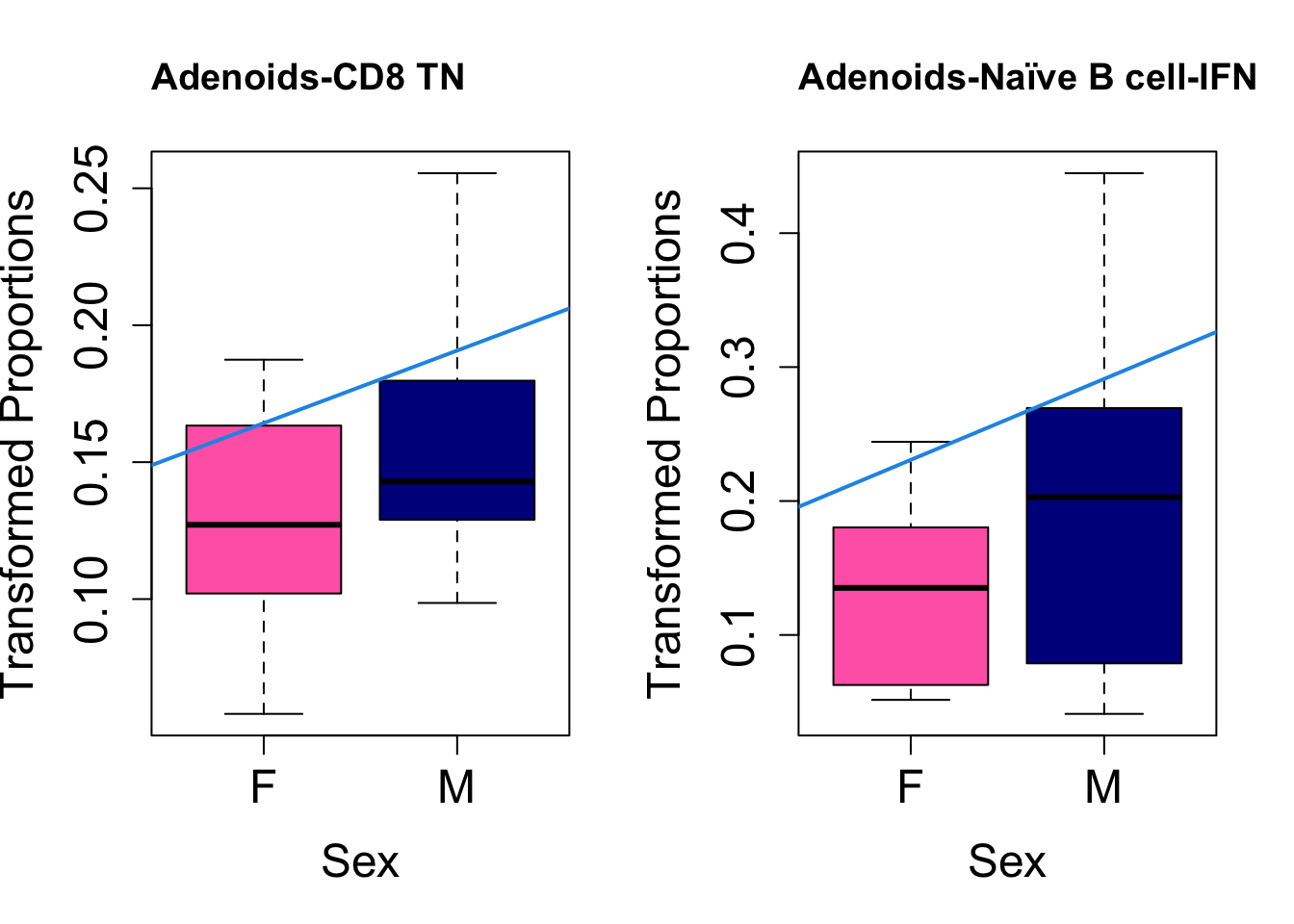
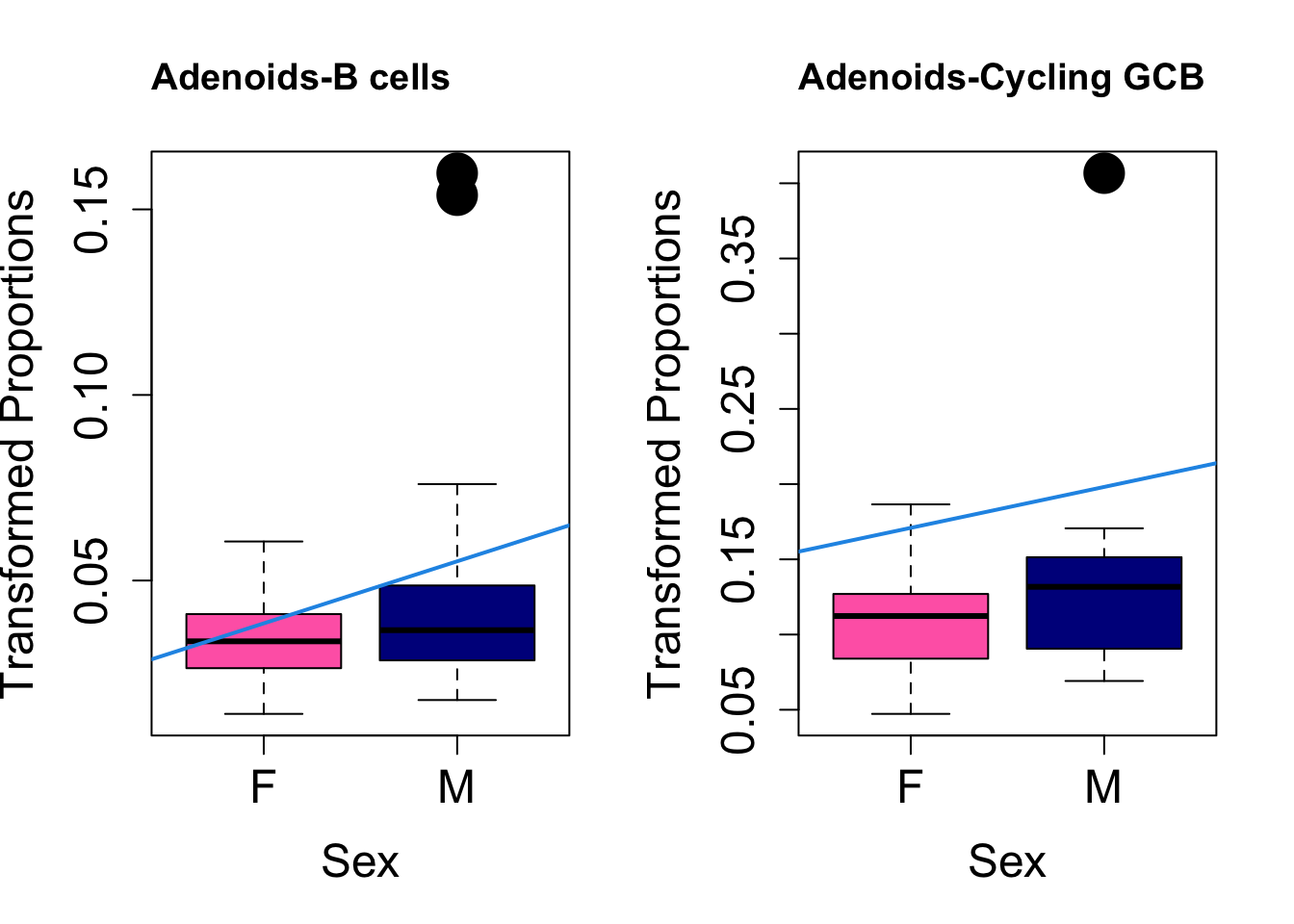
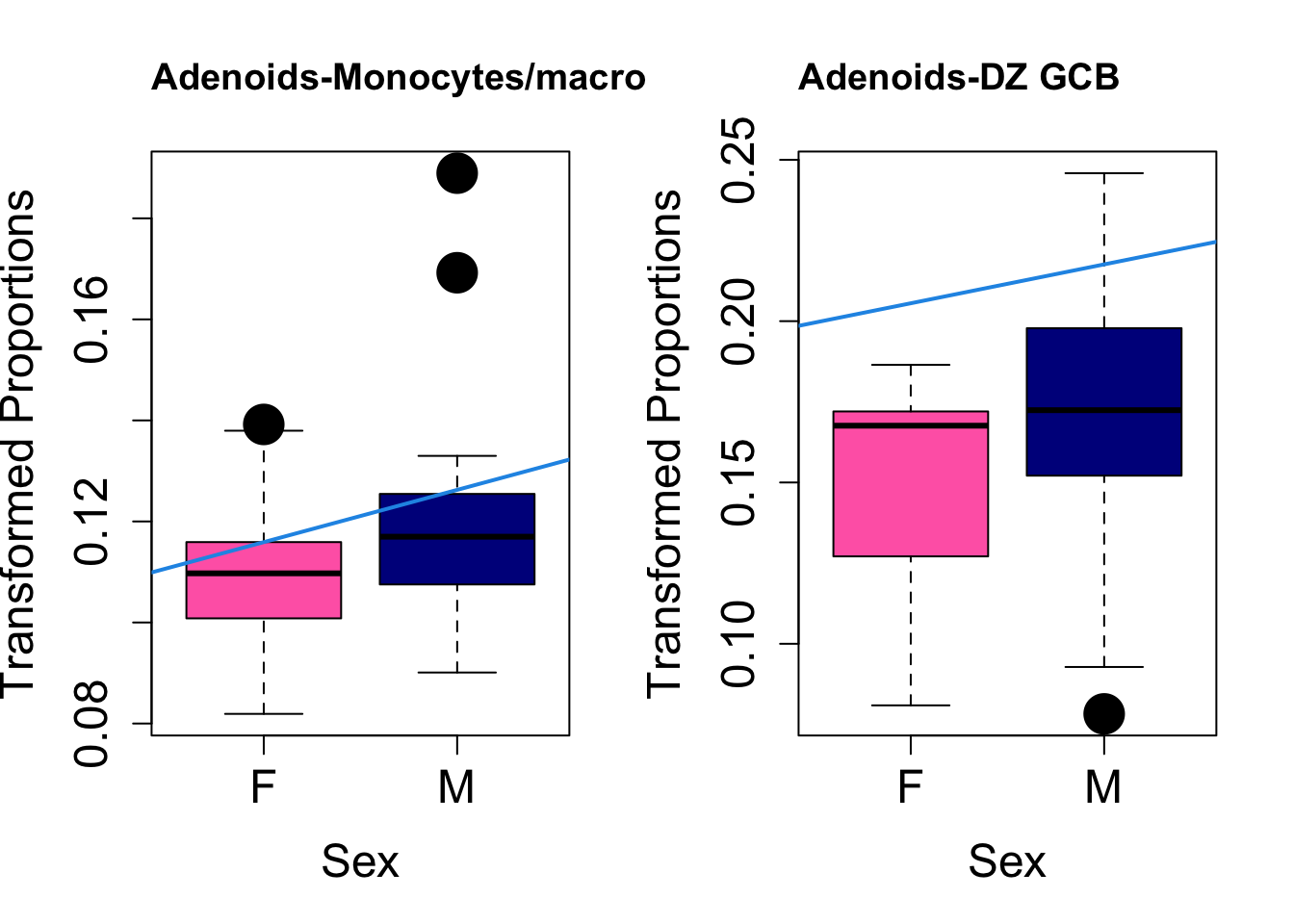
Plot results for Sex
coef = "sexM"
toptable.transformedProps.sex <- topTable(fit, coef = coef)
# Plot Sex
cat(paste0("#### Sex"," {.tabset}\n\n"))#### Sex {.tabset} par(mfrow=c(1,2))
for (i in rownames(toptable.transformedProps.sex)) {
plot(sex, props$TransformedProps[i,],
pch=16, cex=3, ylab="Transformed Proportions", xlab="Sex", cex.lab=1.5, cex.axis=1.5,
cex.main=2, col=c("hotpink", "darkblue"))
abline(a=fit$coefficients[i, 1], b=fit$coefficients[i, 3], col=4,
lwd=2)
title(paste0(tissue, "-", i), cex.main = 1.2, adj = 0)
}
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
Interaction model for Age and Sex
design <- model.matrix(~age*sex)
design (Intercept) age sexM age:sexM
1 1 3.62 1 3.62
2 1 3.29 0 0.00
3 1 6.79 0 0.00
4 1 5.82 0 0.00
5 1 1.64 1 1.64
6 1 3.73 1 3.73
7 1 2.31 1 2.31
8 1 3.82 1 3.82
9 1 6.67 0 0.00
10 1 2.73 1 2.73
11 1 5.02 0 0.00
12 1 3.93 1 3.93
13 1 3.76 1 3.76
14 1 4.45 1 4.45
15 1 5.28 0 0.00
16 1 11.27 0 0.00
17 1 8.90 1 8.90
18 1 15.19 1 15.19
19 1 2.41 1 2.41
20 1 1.58 0 0.00
21 1 2.86 0 0.00
22 1 8.01 1 8.01
23 1 11.16 1 11.16
24 1 7.34 0 0.00
25 1 6.42 1 6.42
26 1 11.57 1 11.57
27 1 10.54 0 0.00
28 1 11.23 1 11.23
29 1 12.75 1 12.75
30 1 7.77 0 0.00
31 1 13.22 0 0.00
32 1 12.21 0 0.00
attr(,"assign")
[1] 0 1 2 3
attr(,"contrasts")
attr(,"contrasts")$sex
[1] "contr.treatment"fit <- lmFit(props$TransformedProps, design)
cont.matrix <- cbind(
AgeInFemales = c(0, 1, 0, 0),
AgeInMales = c(0, 1, 0, 1),
Diff = c(0, 0, 0, 1)
)
cont.matrix AgeInFemales AgeInMales Diff
[1,] 0 0 0
[2,] 1 1 0
[3,] 0 0 0
[4,] 0 1 1fit2 <- contrasts.fit(fit, cont.matrix)
fit2 <- eBayes(fit2)
coef = "Diff"
toptable.transformedProps <- topTable(fit2, coef = coef)
toptable.transformedProps logFC AveExpr t P.Value adj.P.Val
Neutrophils -0.008238283 0.02632626 -2.882840 0.007139463 0.2641601
Plasma B cells -0.006036302 0.16304236 -2.267821 0.030538199 0.5649567
csMBC FCRL4/5+ 0.002310903 0.07141775 1.742044 0.091523758 0.5853223
B cells 0.004615980 0.04415960 1.587945 0.122557403 0.5853223
CD8 TN 0.006390036 0.14835309 1.584799 0.123271556 0.5853223
CD8 TF 0.004598912 0.15471351 1.581915 0.123929182 0.5853223
CD4 TCM 0.005191868 0.13350483 1.508555 0.141647298 0.5853223
Double negative T 0.002530382 0.06317316 1.481409 0.148700545 0.5853223
CD4 T proliferating 0.002691442 0.07685101 1.456340 0.155461715 0.5853223
Monocytes/macrophages 0.003007789 0.11595110 1.439594 0.160113095 0.5853223
B
Neutrophils -4.480896
Plasma B cells -5.821961
csMBC FCRL4/5+ -6.782023
B cells -7.024793
CD8 TN -7.029552
CD8 TF -7.033907
CD4 TCM -7.142429
Double negative T -7.181460
CD4 T proliferating -7.216961
Monocytes/macrophages -7.240381Save results
out <- here("output",
"CSV_v2",
paste0("G000231_Neeland_",tissue,".propeller.xlsx"))
if (!file.exists(out)) {
write.xlsx(toptable.transformedProps, file = out, rowNames= T)
}Age modelling across GC cell subsets (population)
idx <- which(merged_obj$cell_labels %in% "Germinal centre B cells")
merged_obj <- merged_obj[,idx]
merged_objAn object of class Seurat
17456 features across 42615 samples within 1 assay
Active assay: RNA (17456 features, 2000 variable features)
4 layers present: data.2, counts.2, scale.data.2, scale.data
2 dimensional reductions calculated: pca, umap.mergedRepeat modeling steps
metadata_df <- data.frame(
sample = merged_obj$sample_id,
#donor = merged_obj$donor_id,
age_years = as.character(merged_obj$age_years),
cell_type = merged_obj$cell_labels_v2
)
metadata_df$age_years <- as.numeric(metadata_df$age_years)
# Calculate cell type proportions within each sample, age group, and cell type
barplot_data <- metadata_df %>%
group_by(sample, age_years, cell_type) %>%
summarise(n_cells = n()) %>%
ungroup() %>%
group_by(sample, age_years) %>%
mutate(n_cells_total = sum(n_cells)) %>%
ungroup() %>%
mutate(percentage_cells = n_cells / n_cells_total)`summarise()` has grouped output by 'sample', 'age_years'. You can override
using the `.groups` argument.barplot_data <- barplot_data %>%
arrange(age_years)
b <- ggplot(barplot_data, aes(x = reorder(paste(sample, age_years, sep = ":"), age_years),
y = percentage_cells, fill = cell_type)) +
geom_bar(stat = "identity") +
ggtitle(paste0("Age vs Cell Type Proportions (T cell population): ", tissue)) +
labs(x = "Sample:Age (Years)", y = "Proportion", fill = "Cell Type") +
scale_fill_manual(values = color_palette) +
theme_minimal() +
theme(
plot.title = element_text(size = 13, hjust = 0.5, face = "bold"),
legend.position = "top",
axis.text.x = element_text(angle = 45, hjust = 1)
)
b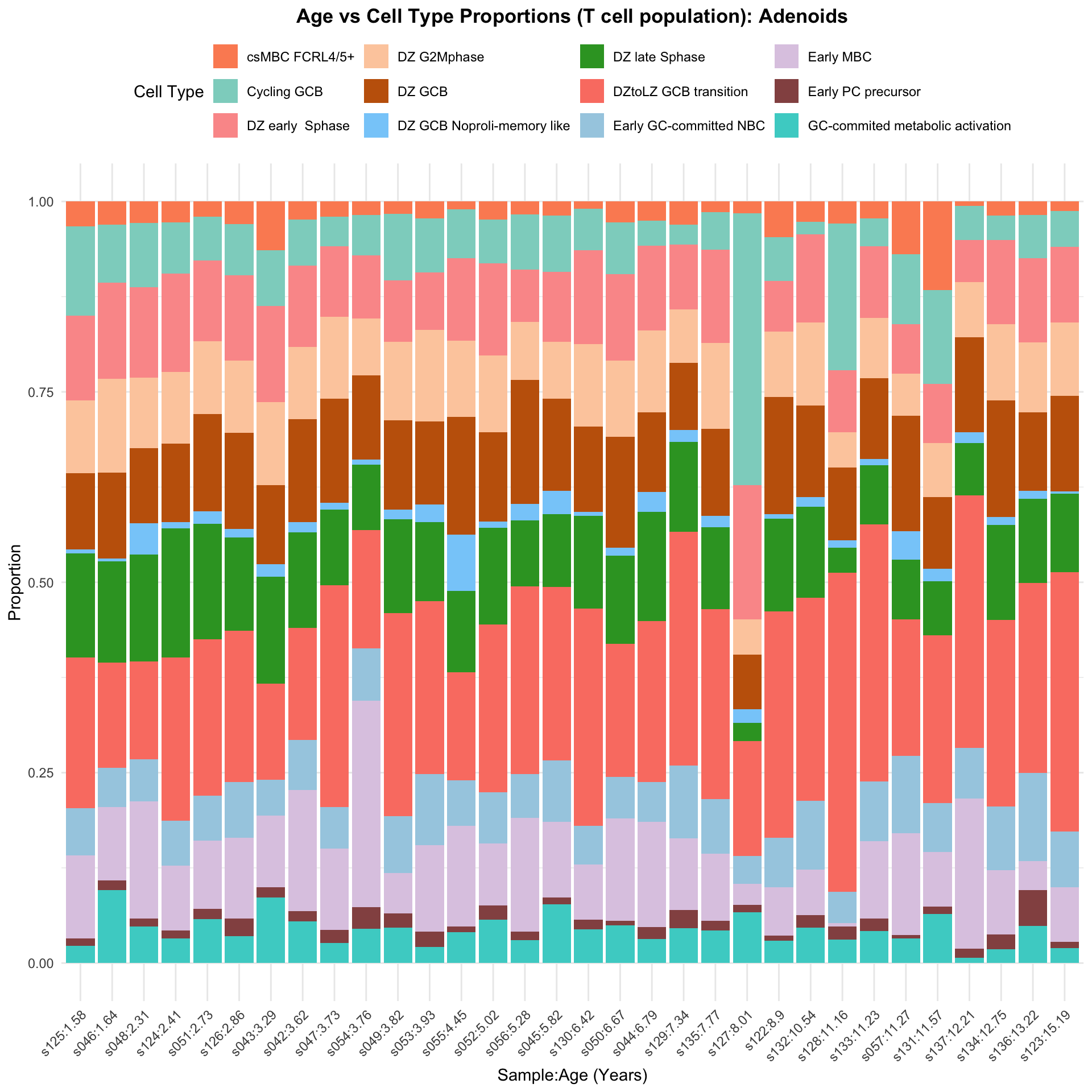
props <- getTransformedProps(clusters = merged_obj$cell_labels_v2,
sample = merged_obj$sample_id, transform = "asin")Performing arcsin square root transformation of proportions# Plot Cell Type Mean Variance
p1 <- plotCellTypeMeanVar(props$Counts)Using classic mode.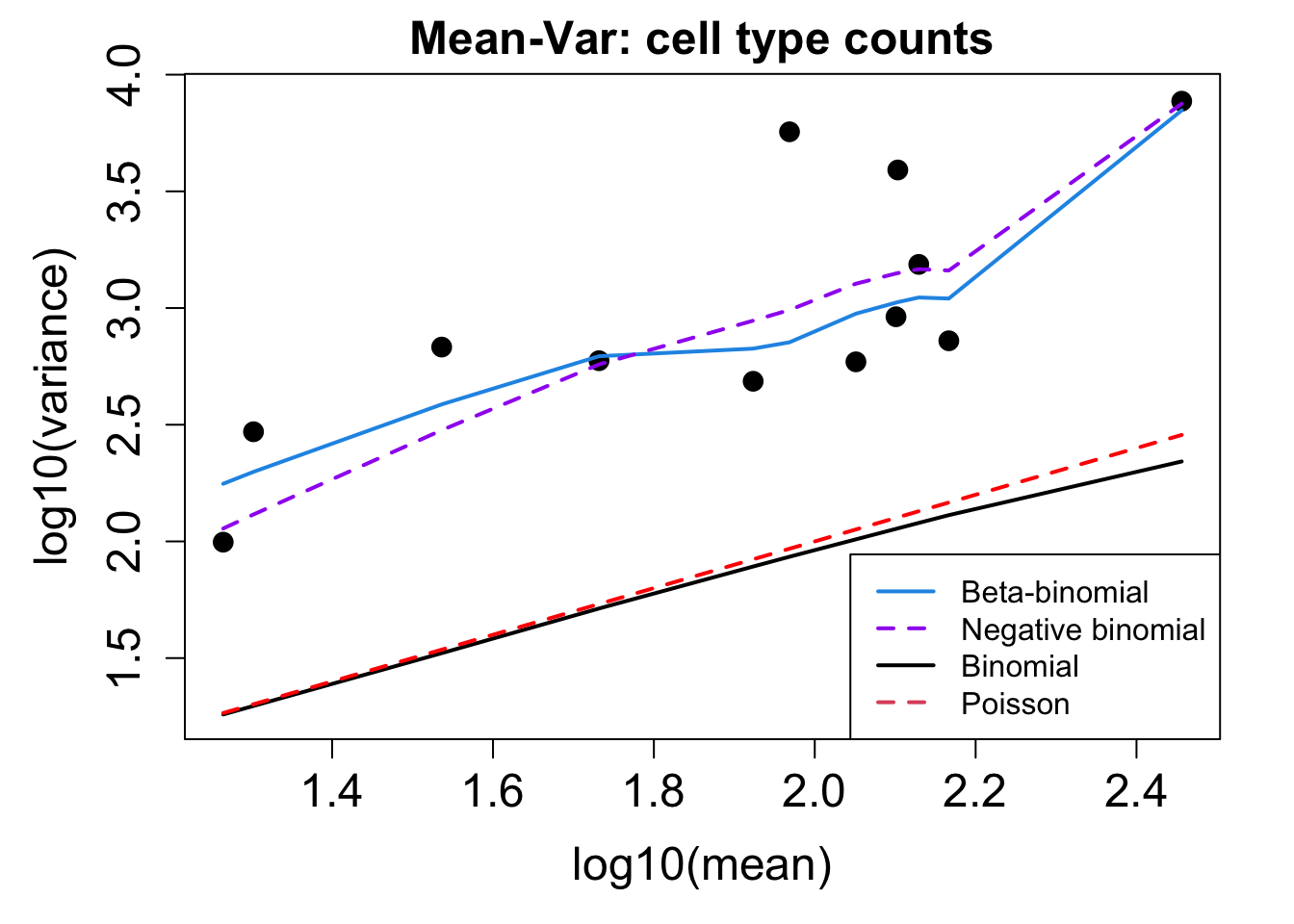
# Plot Cell Type Proportions Mean Variance
p2 <- plotCellTypePropsMeanVar(props$Counts)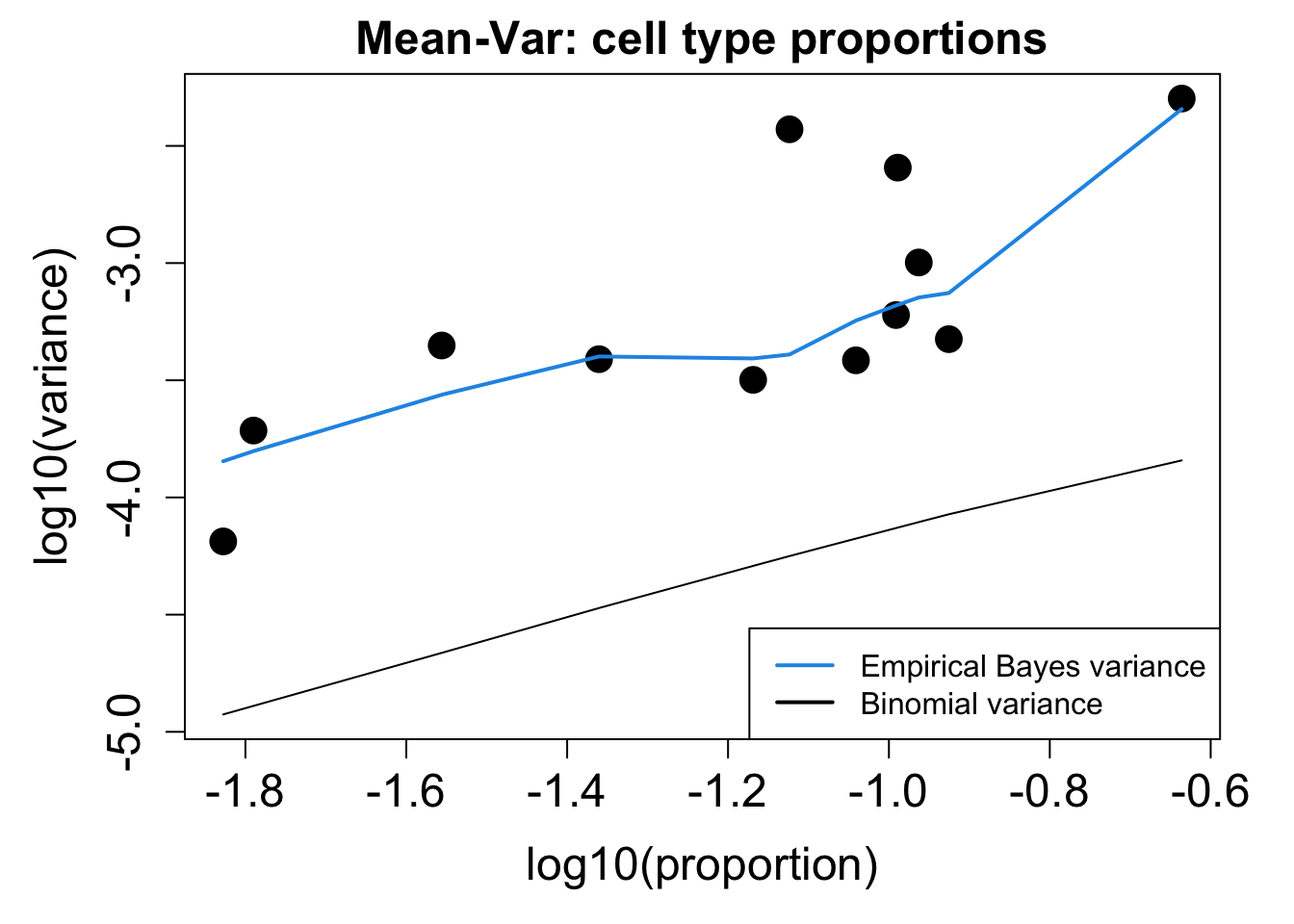
p1 / p2numeric(0)print(knitr::kable(props$Proportions, caption = "Cell-type proportions (GC cell population) in samples"))
Table: Cell-type proportions (GC cell population) in samples
| | s042| s043| s044| s045| s046| s047| s048| s049| s050| s051| s052| s053| s054| s055| s056| s057| s122| s123| s124| s125| s126| s127| s128| s129| s130| s131| s132| s133| s134| s135| s136| s137|
|:--------------------------------|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|---------:|
|csMBC FCRL4/5+ | 0.0240700| 0.0637119| 0.0249671| 0.0186386| 0.0302267| 0.0202775| 0.0284957| 0.0162602| 0.0274415| 0.0198864| 0.0234474| 0.0222822| 0.0176668| 0.0105798| 0.0168350| 0.0691244| 0.0466563| 0.0124138| 0.0276119| 0.0326087| 0.0293564| 0.0156250| 0.0289608| 0.0307167| 0.0094406| 0.1165049| 0.0267335| 0.0224467| 0.0188867| 0.0138504| 0.0178947| 0.0058939|
|Cycling GCB | 0.0598104| 0.0734072| 0.0328515| 0.0737439| 0.0764064| 0.0384205| 0.0841617| 0.0872136| 0.0677966| 0.0577652| 0.0576679| 0.0708980| 0.0533049| 0.0639018| 0.0729517| 0.0921659| 0.0575428| 0.0468966| 0.0671642| 0.1173913| 0.0677456| 0.3571429| 0.1925043| 0.0255973| 0.0545455| 0.1229773| 0.0167084| 0.0364759| 0.0318091| 0.0493075| 0.0568421| 0.0445318|
|DZ early Sphase | 0.1072210| 0.1260388| 0.1116951| 0.0915721| 0.1263644| 0.0928495| 0.1186216| 0.0805617| 0.1138015| 0.1060606| 0.1210393| 0.0756246| 0.0828511| 0.1083369| 0.0684624| 0.0645161| 0.0668740| 0.0993103| 0.1291045| 0.1108696| 0.1117802| 0.1755952| 0.0817717| 0.0853242| 0.1230769| 0.0776699| 0.1152882| 0.0942761| 0.1103380| 0.1224377| 0.1105263| 0.0556647|
|DZ G2Mphase | 0.0948213| 0.1094183| 0.1077530| 0.0753647| 0.1234257| 0.1077908| 0.0927767| 0.1034738| 0.1000807| 0.0956439| 0.1007605| 0.1201891| 0.0743223| 0.0998730| 0.0763187| 0.0552995| 0.0855365| 0.0965517| 0.0940299| 0.0956522| 0.0948438| 0.0468750| 0.0459966| 0.0699659| 0.1087413| 0.0711974| 0.1094403| 0.0785634| 0.1003976| 0.1130194| 0.0915789| 0.0720367|
|DZ GCB | 0.1349380| 0.1038781| 0.1038108| 0.1207455| 0.1120907| 0.1366062| 0.0987409| 0.1167775| 0.1452785| 0.1278409| 0.1172370| 0.1087103| 0.1108742| 0.1544647| 0.1627385| 0.1520737| 0.1539658| 0.1255172| 0.1029851| 0.1000000| 0.1260820| 0.0714286| 0.0954003| 0.0887372| 0.1115385| 0.0938511| 0.1203008| 0.1066218| 0.1530815| 0.1141274| 0.1031579| 0.1250819|
|DZ GCB Noproli-memory like | 0.0138585| 0.0166205| 0.0262812| 0.0307942| 0.0037783| 0.0085379| 0.0404241| 0.0133038| 0.0104923| 0.0160985| 0.0082383| 0.0236327| 0.0063966| 0.0740584| 0.0213244| 0.0368664| 0.0062208| 0.0027586| 0.0082090| 0.0054348| 0.0112909| 0.0178571| 0.0102215| 0.0153584| 0.0052448| 0.0161812| 0.0125313| 0.0078563| 0.0099404| 0.0149584| 0.0105263| 0.0137525|
|DZ late Sphase | 0.1254559| 0.1398892| 0.1432326| 0.0956240| 0.1335013| 0.0992529| 0.1404904| 0.1226903| 0.1162228| 0.1515152| 0.1267427| 0.1033086| 0.0862016| 0.1066441| 0.0864198| 0.0783410| 0.1213064| 0.1034483| 0.1694030| 0.1369565| 0.1223184| 0.0238095| 0.0323680| 0.1177474| 0.1220280| 0.0711974| 0.1194653| 0.0780022| 0.1252485| 0.1074792| 0.1105263| 0.0687623|
|DZtoLZ GCB transition | 0.1466083| 0.1260388| 0.2115637| 0.2277147| 0.1381192| 0.2913554| 0.1285620| 0.2668145| 0.1743341| 0.2054924| 0.2205323| 0.2268737| 0.1553457| 0.1421921| 0.2469136| 0.1797235| 0.2970451| 0.3406897| 0.2149254| 0.1978261| 0.1987204| 0.1510417| 0.4190801| 0.3071672| 0.2849650| 0.2200647| 0.2664996| 0.3372615| 0.2445328| 0.2493075| 0.2494737| 0.3320236|
|Early GC-committed NBC | 0.0656455| 0.0470914| 0.0525624| 0.0802269| 0.0512175| 0.0544290| 0.0556660| 0.0746489| 0.0548830| 0.0587121| 0.0671736| 0.0938555| 0.0682303| 0.0596699| 0.0572391| 0.1013825| 0.0653188| 0.0731034| 0.0589552| 0.0619565| 0.0730147| 0.0364583| 0.0408859| 0.0955631| 0.0506993| 0.0647249| 0.0902256| 0.0785634| 0.0834990| 0.0720222| 0.1157895| 0.0661428|
|Early MBC | 0.1590080| 0.0941828| 0.1379763| 0.0996759| 0.0965575| 0.1067236| 0.1537442| 0.0532151| 0.1339790| 0.0899621| 0.0817490| 0.1134369| 0.2713981| 0.1320355| 0.1492705| 0.1336406| 0.0637636| 0.0717241| 0.0850746| 0.1086957| 0.1065111| 0.0275298| 0.0051107| 0.0938567| 0.0730769| 0.0711974| 0.0601504| 0.1015713| 0.0844930| 0.0880886| 0.0378947| 0.1971185|
|Early PC precursor | 0.0138585| 0.0138504| 0.0157687| 0.0089141| 0.0125945| 0.0170758| 0.0106030| 0.0184775| 0.0064568| 0.0132576| 0.0183777| 0.0202566| 0.0280231| 0.0080406| 0.0112233| 0.0046083| 0.0062208| 0.0082759| 0.0104478| 0.0097826| 0.0233346| 0.0096726| 0.0170358| 0.0238908| 0.0122378| 0.0097087| 0.0158730| 0.0162738| 0.0198807| 0.0127424| 0.0473684| 0.0117878|
|GC-commited metabolic activation | 0.0547046| 0.0858726| 0.0315375| 0.0769854| 0.0957179| 0.0266809| 0.0477137| 0.0465632| 0.0492333| 0.0577652| 0.0570342| 0.0209318| 0.0453853| 0.0402031| 0.0303030| 0.0322581| 0.0295490| 0.0193103| 0.0320896| 0.0228261| 0.0350019| 0.0669643| 0.0306644| 0.0460751| 0.0444056| 0.0647249| 0.0467836| 0.0420875| 0.0178926| 0.0426593| 0.0484211| 0.0072037|fit <- lmFit(props$TransformedProps, design)
fit <- eBayes(fit, robust=TRUE)
toptable.transformedProps <- topTable(fit) Removing intercept from test coefficientsfit.prop <- lmFit(props$Proportions, design)
fit.prop <- eBayes(fit.prop, robust=TRUE)
toptable.props <- topTable(fit.prop, sort.by = "F")Removing intercept from test coefficientsprint(knitr::kable(toptable.transformedProps, caption = paste0("Transformed proportions Toptable results: ", tissue)))
Table: Transformed proportions Toptable results: Adenoids
| | age| sexM| age.sexM| AveExpr| F| P.Value| adj.P.Val|
|:--------------------------------|----------:|----------:|----------:|---------:|---------:|---------:|---------:|
|DZtoLZ GCB transition | 0.0094888| -0.0239776| 0.0051743| 0.4973090| 5.9046469| 0.0024982| 0.0299789|
|Early GC-committed NBC | 0.0068340| 0.0204215| -0.0057300| 0.2614005| 3.5566912| 0.0246152| 0.1229153|
|DZ late Sphase | -0.0059952| -0.0053302| -0.0018078| 0.3321544| 3.3460571| 0.0307288| 0.1229153|
|Early MBC | -0.0018975| 0.0322420| -0.0091148| 0.3162679| 2.3354358| 0.0922760| 0.2768279|
|DZ G2Mphase | -0.0029690| 0.0048775| -0.0010462| 0.3046049| 1.6142842| 0.2047316| 0.4913557|
|GC-commited metabolic activation | -0.0040201| 0.0015062| -0.0004909| 0.2051218| 1.3039716| 0.2895196| 0.5530865|
|Cycling GCB | -0.0072293| -0.0260940| 0.0095590| 0.2656987| 1.0147792| 0.3989096| 0.5530865|
|DZ GCB Noproli-memory like | 0.0022498| 0.0300034| -0.0060151| 0.1198400| 0.9791156| 0.4143870| 0.5530865|
|DZ early Sphase | -0.0047578| -0.0173860| 0.0028707| 0.3230403| 0.9781693| 0.4148149| 0.5530865|
|csMBC FCRL4/5+ | -0.0035594| -0.0487736| 0.0061295| 0.1598554| 0.5436824| 0.6558179| 0.7869815|print(knitr::kable(toptable.props, caption = paste0("Proportions Toptable results: ", tissue)))
Table: Proportions Toptable results: Adenoids
| | age| sexM| age.sexM| AveExpr| F| P.Value| adj.P.Val|
|:--------------------------------|----------:|----------:|----------:|---------:|---------:|---------:|---------:|
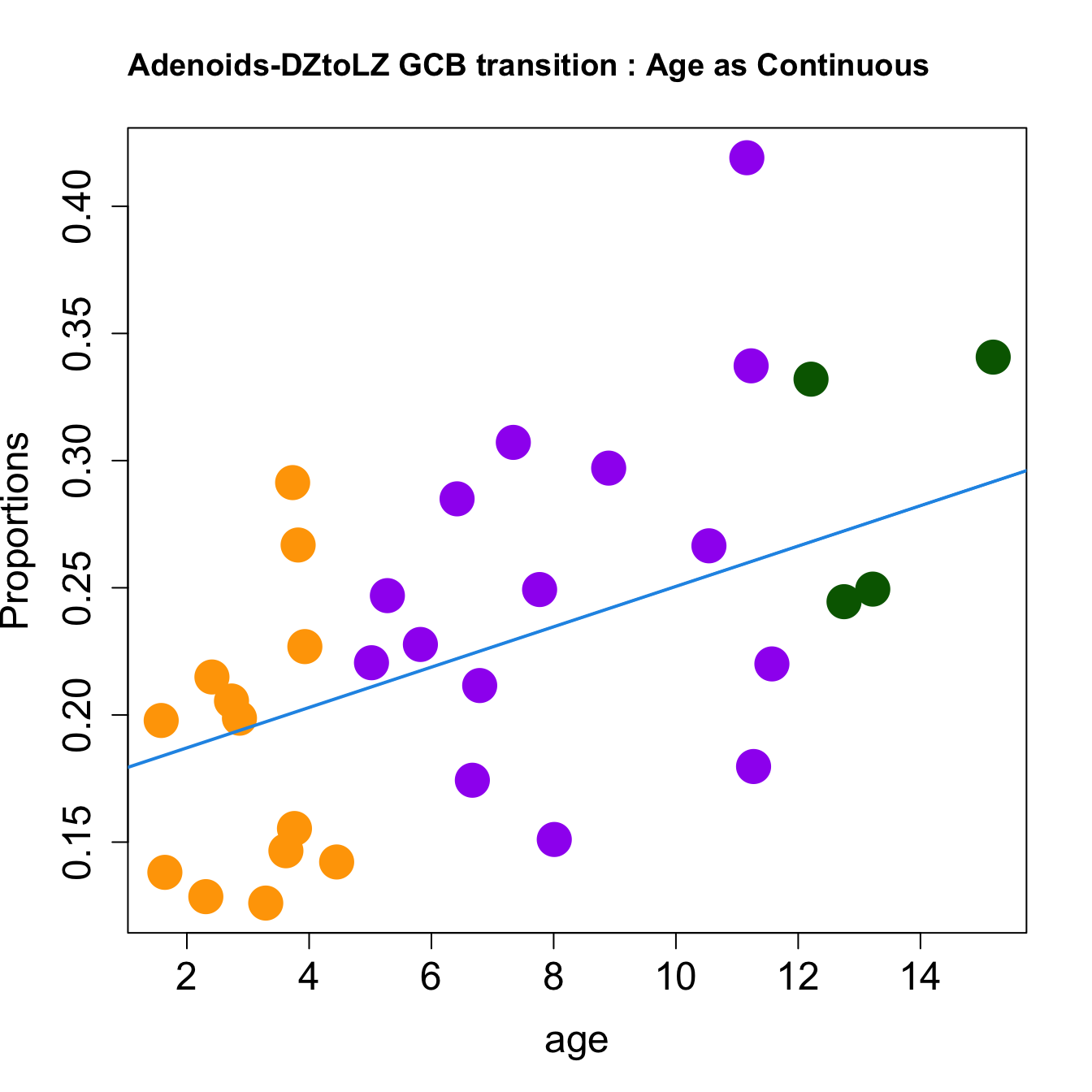
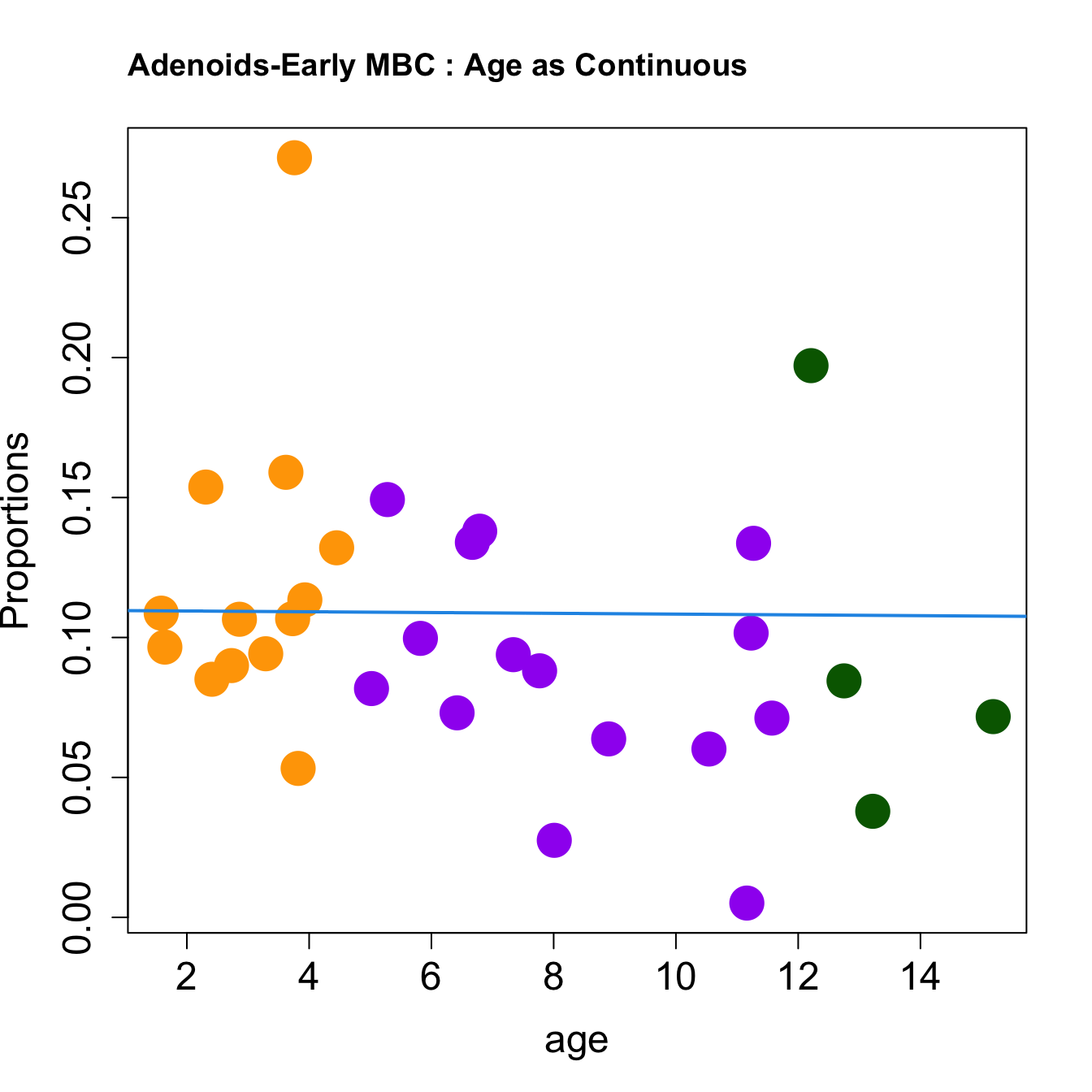
|DZtoLZ GCB transition | 0.0079384| -0.0187380| 0.0045311| 0.2312128| 5.8959280| 0.0027560| 0.0330723|
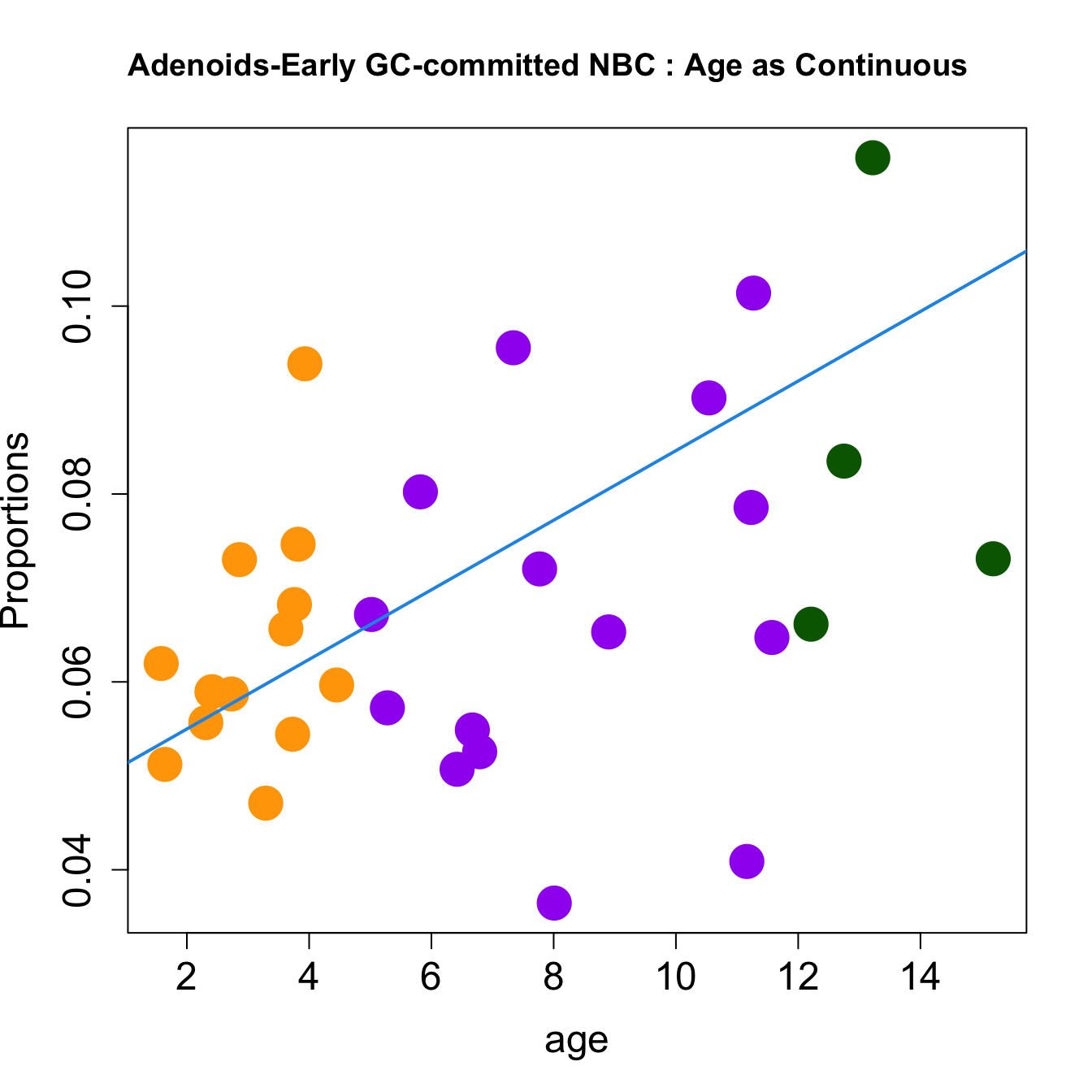
|Early GC-committed NBC | 0.0037044| 0.0115831| -0.0031146| 0.0677986| 4.5020169| 0.0100096| 0.0600578|
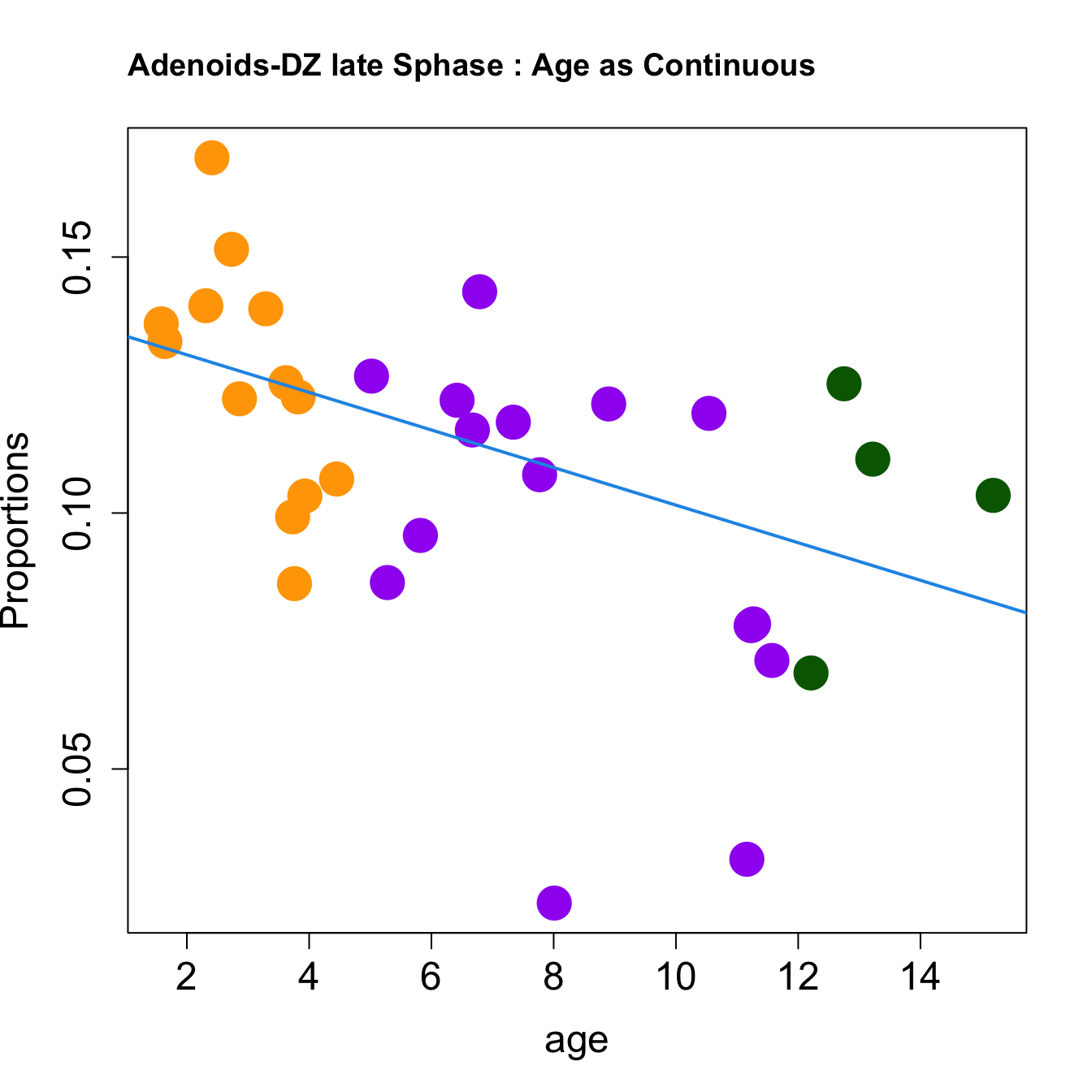
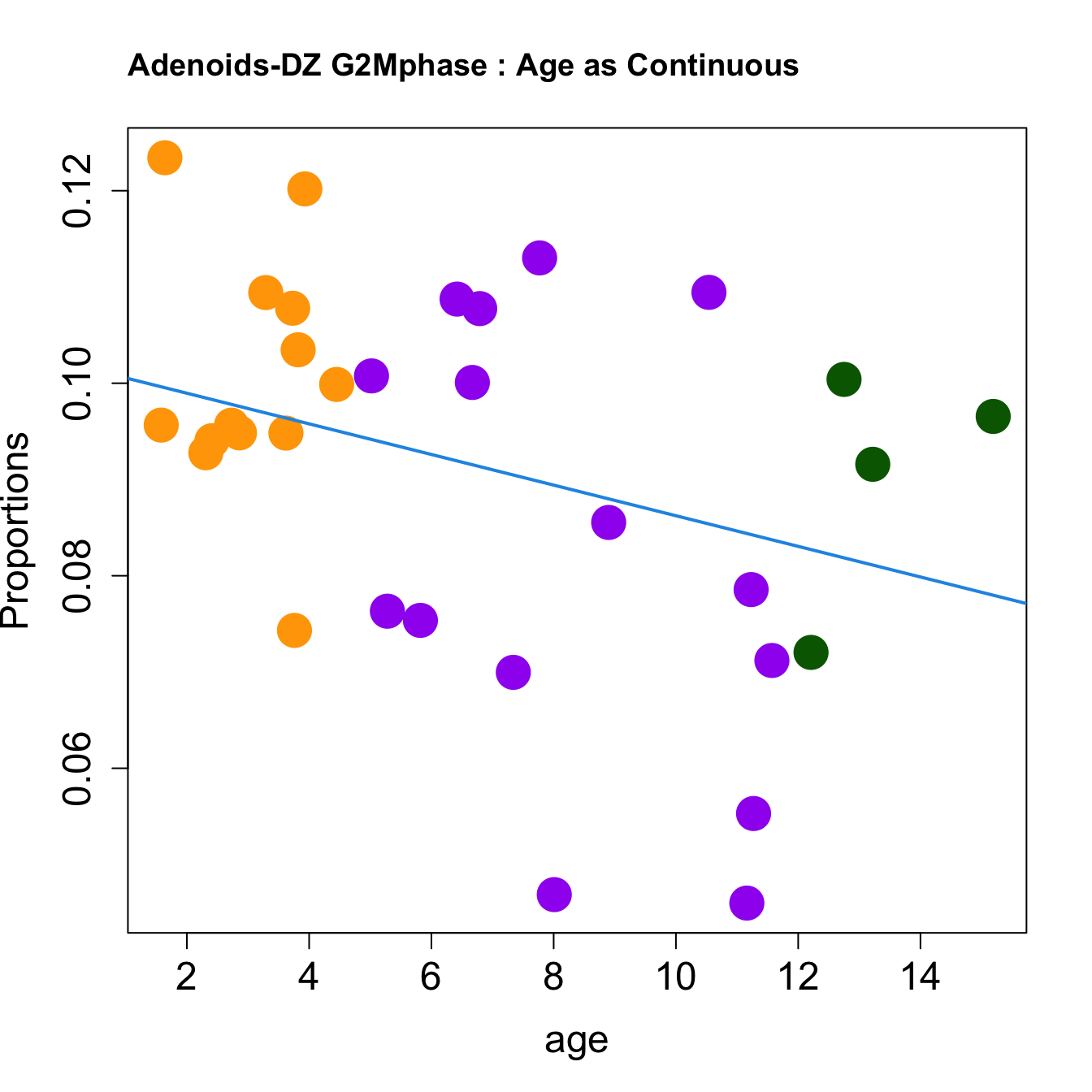
|DZ late Sphase | -0.0036712| -0.0024719| -0.0008197| 0.1089250| 3.7246549| 0.0216838| 0.0867354|
|Early MBC | -0.0001415| 0.0276792| -0.0059335| 0.1025754| 1.8019465| 0.1682052| 0.4182741|
|DZ G2Mphase | -0.0015913| 0.0034211| -0.0006199| 0.0909918| 1.7682361| 0.1742809| 0.4182741|
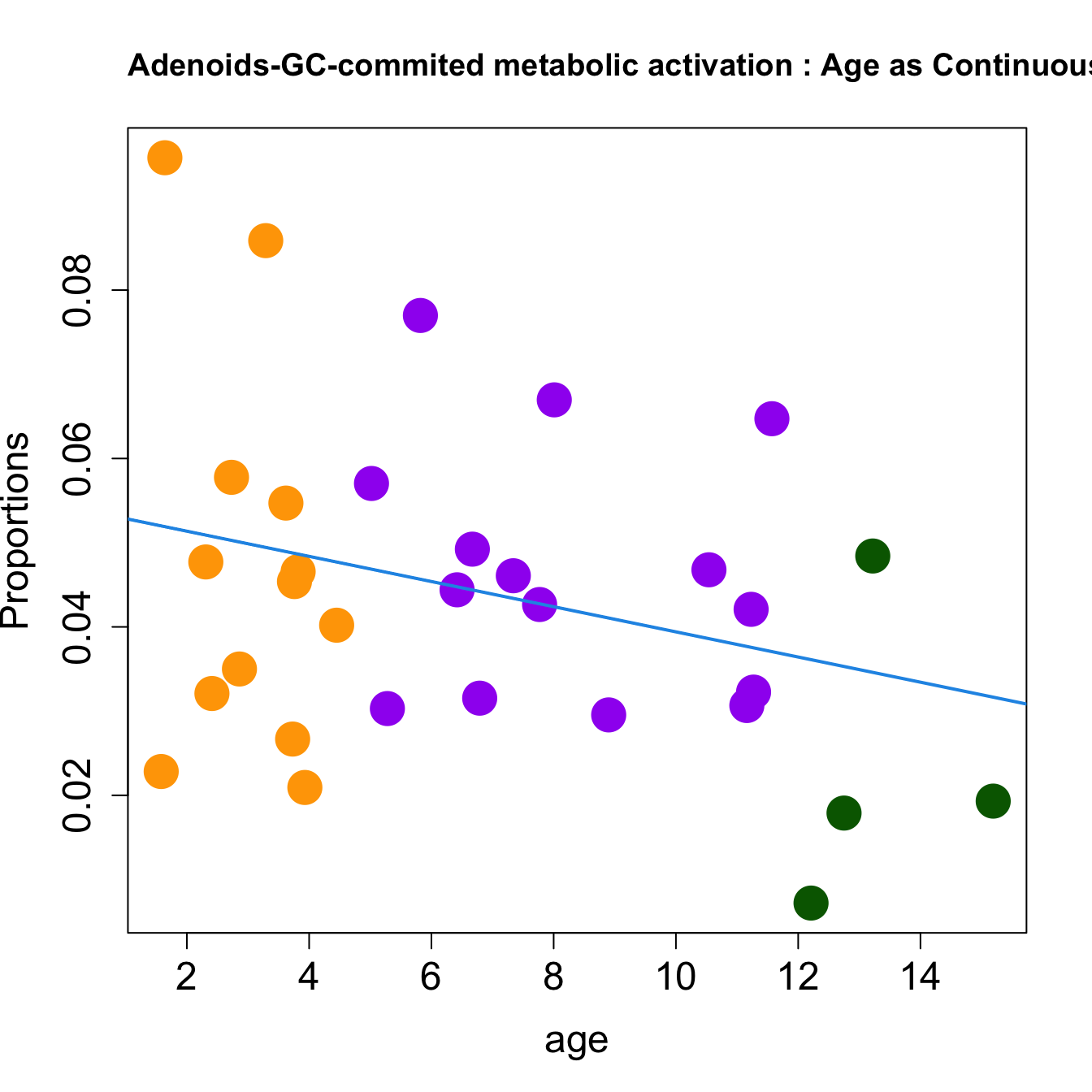
|GC-commited metabolic activation | -0.0014956| 0.0006809| -0.0002697| 0.0436109| 1.1459122| 0.3464400| 0.6928801|
|DZ early Sphase | -0.0026563| -0.0091485| 0.0015446| 0.1020476| 0.8983355| 0.4533959| 0.7206013|
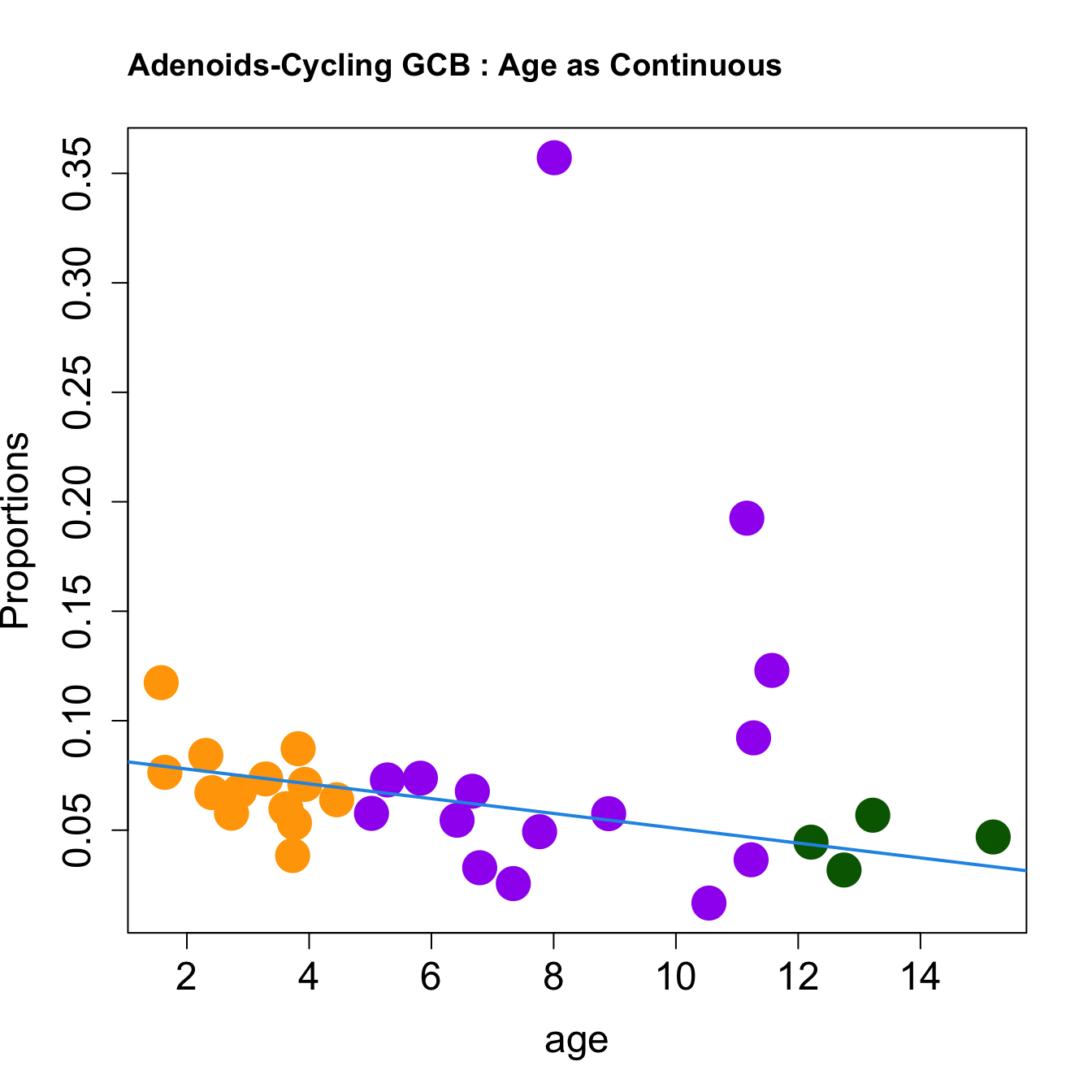
|Cycling GCB | -0.0033816| -0.0136728| 0.0057667| 0.0752391| 0.8316456| 0.4870624| 0.7206013|
|Early PC precursor | 0.0009084| 0.0057230| -0.0011009| 0.0148725| 0.6578154| 0.5843738| 0.7206013|
|DZ GCB Noproli-memory like | 0.0005529| 0.0099786| -0.0016211| 0.0162203| 0.6313186| 0.6005011| 0.7206013|get_age_group_color <- function(age) {
if (age >= 1 && age <= 5) {
return("orange") # Preschool (1-5 years)
} else if (age > 5 && age <= 12) {
return("purple") # Kids (6-11 years)
} else if (age > 12 && age <= 17) {
return("darkgreen") # Adolescent (12-17 years)
} else {
return("black") # Default color for other cases
}
}
age_group_colors <- sapply(age, get_age_group_color)
sorted_indices <- match(rownames(toptable.transformedProps), rownames(props$Proportions))
par(mfrow=c(1,1))
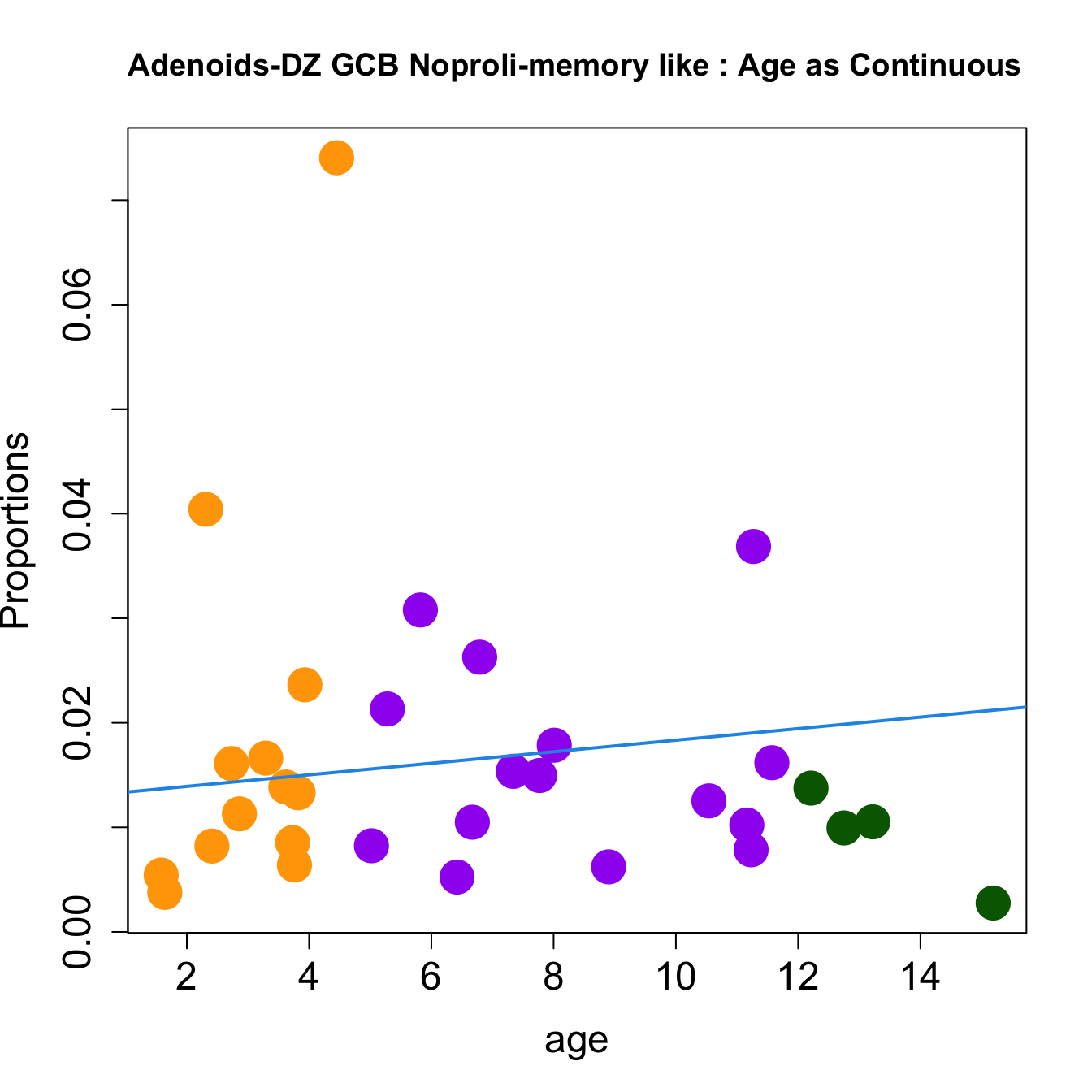
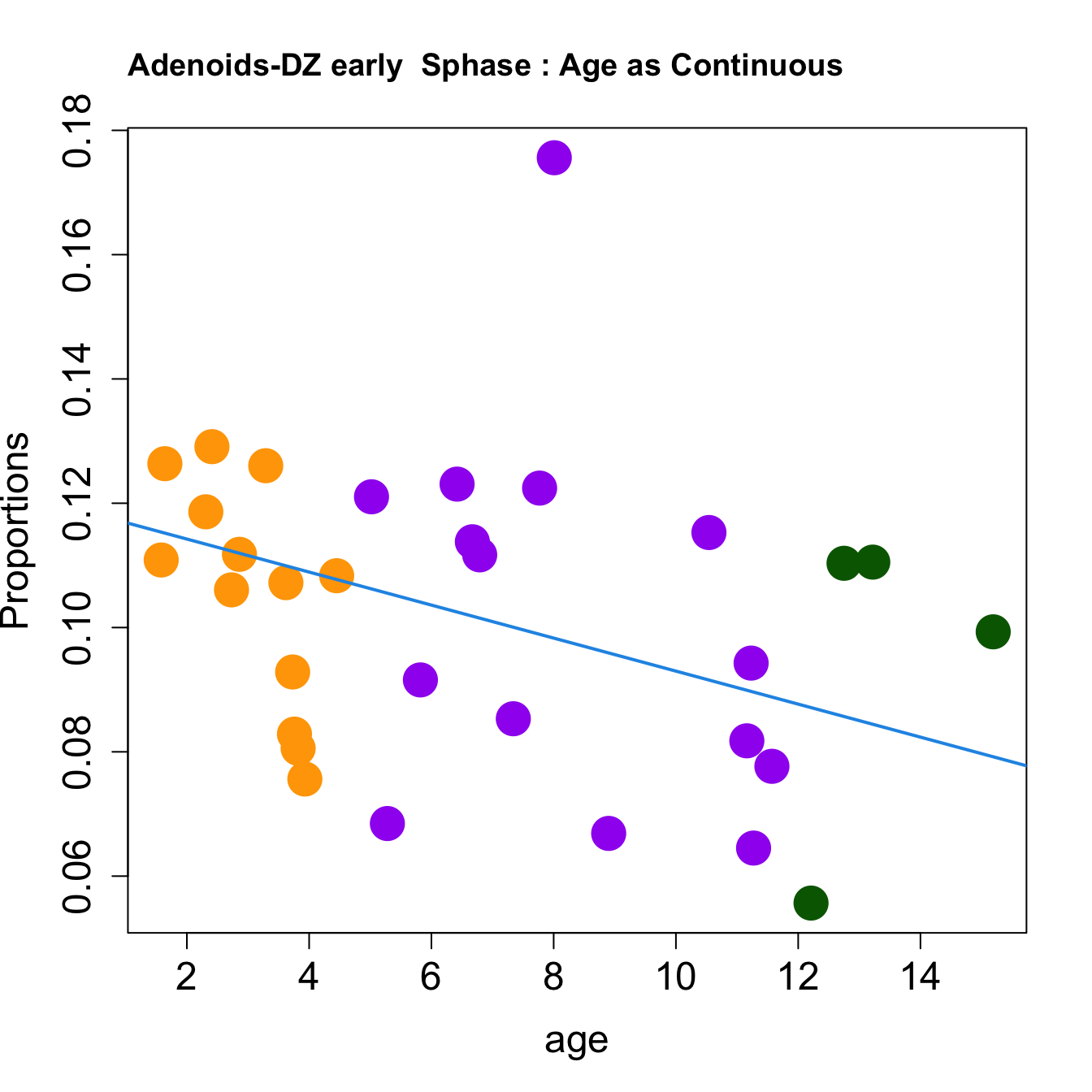
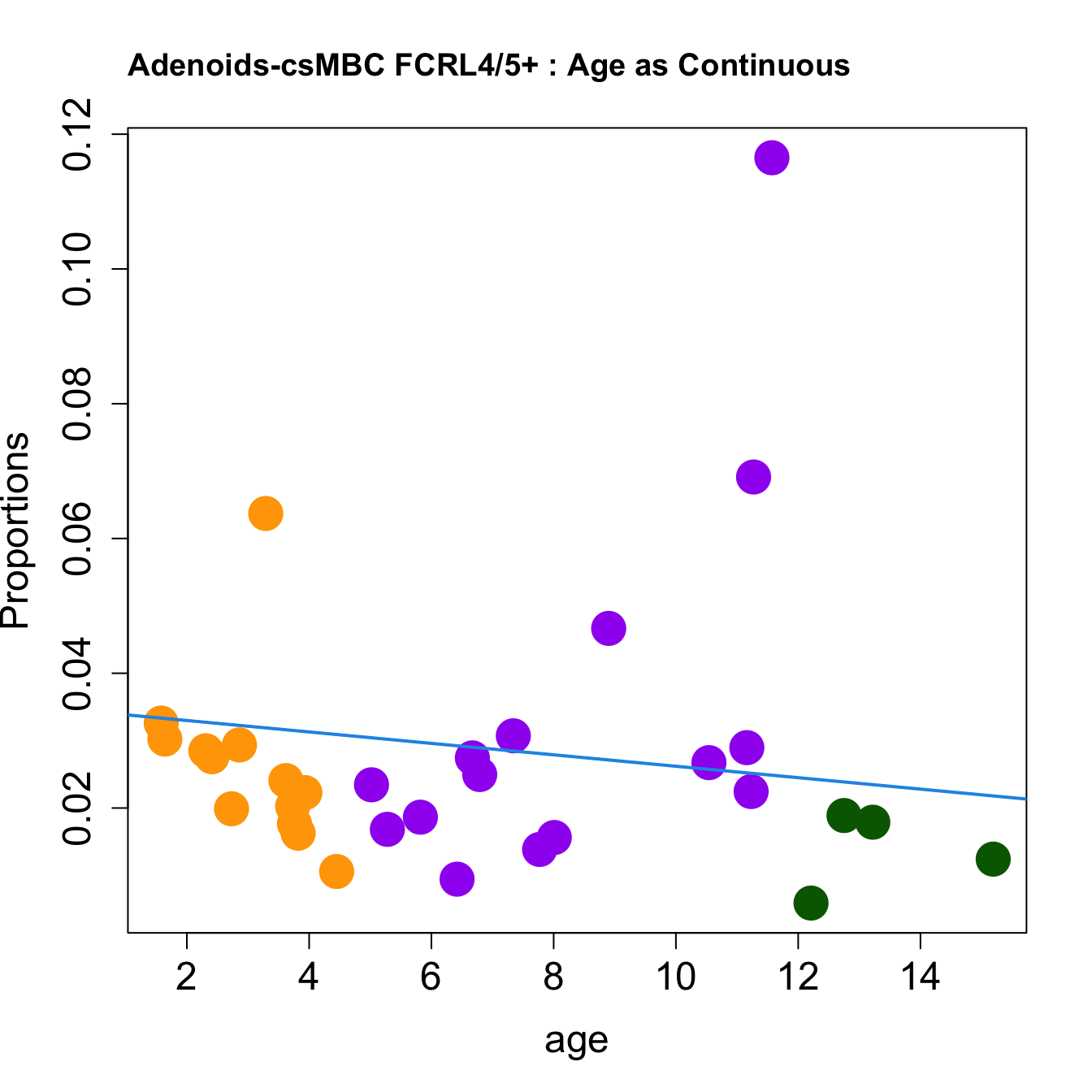
for (i in sorted_indices) {
plot(age, props$Proportions[i,],
pch=16, cex=3, ylab="Proportions", cex.lab=1.5, cex.axis=1.5,cex.main=2, col=age_group_colors)
abline(a=fit.prop$coefficients[i, 1], b=fit.prop$coefficients[i, 2], col=4, lwd=2)
title(paste0(tissue, "-", rownames(props$Proportions)[i], " : Age as Continuous"), cex.main = 1.2, adj = 0)
}
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
| Version | Author | Date |
|---|---|---|
| 0f07f72 | Gunjan Dixit | 2024-10-07 |
# Plot Sex
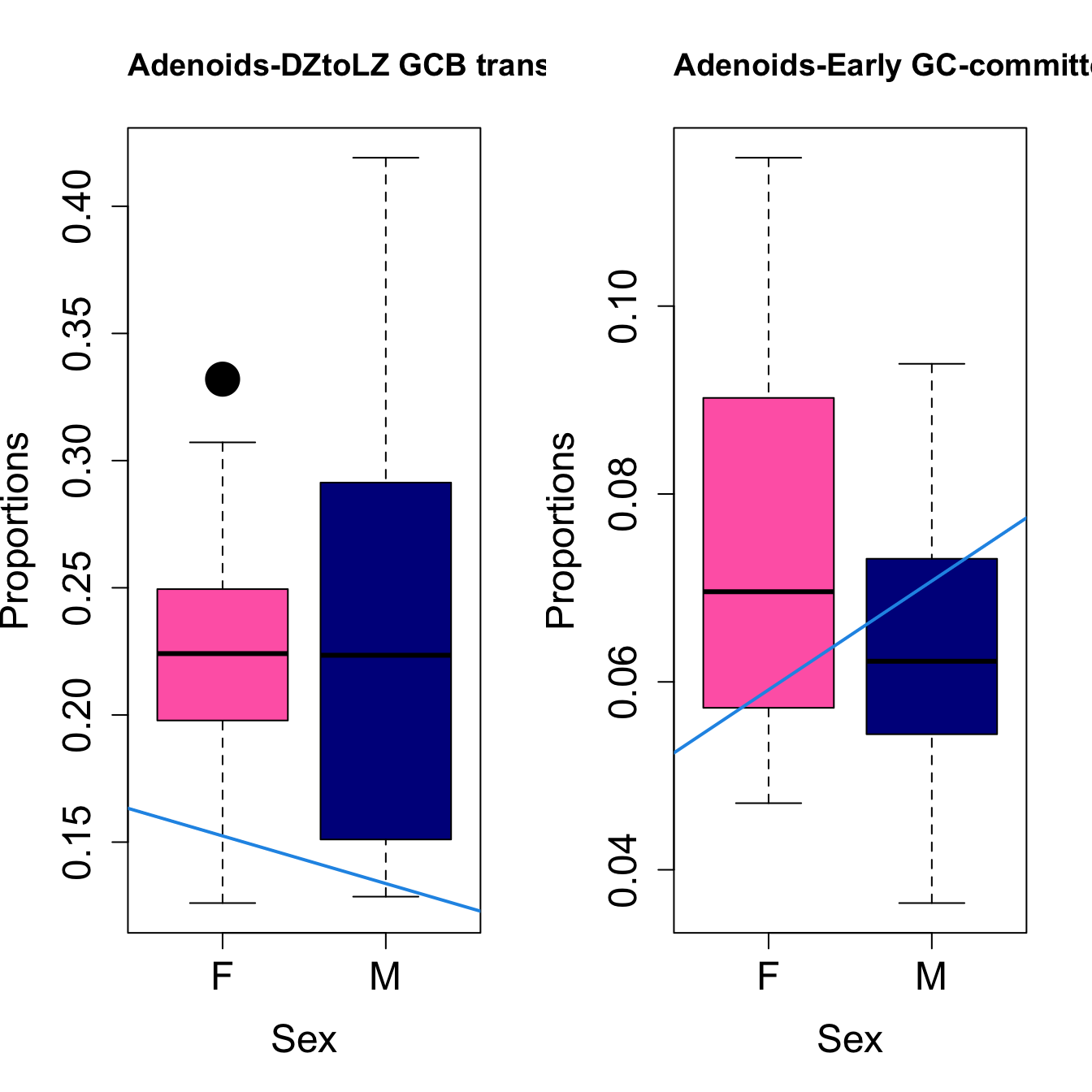
cat(paste0("#### Sex"," {.tabset}\n\n"))#### Sex {.tabset} par(mfrow=c(1,2))
for (i in sorted_indices) {
plot(sex, props$Proportions[i,],
pch=16, cex=3, ylab="Proportions", xlab="Sex", cex.lab=1.5, cex.axis=1.5,
cex.main=2, col=c("hotpink", "darkblue"))
abline(a=fit.prop$coefficients[i, 1], b=fit.prop$coefficients[i, 3], col=4,
lwd=2)
title(paste0(tissue, "-", rownames(props$Proportions)[i]), cex.main = 1.2, adj = 0)
}
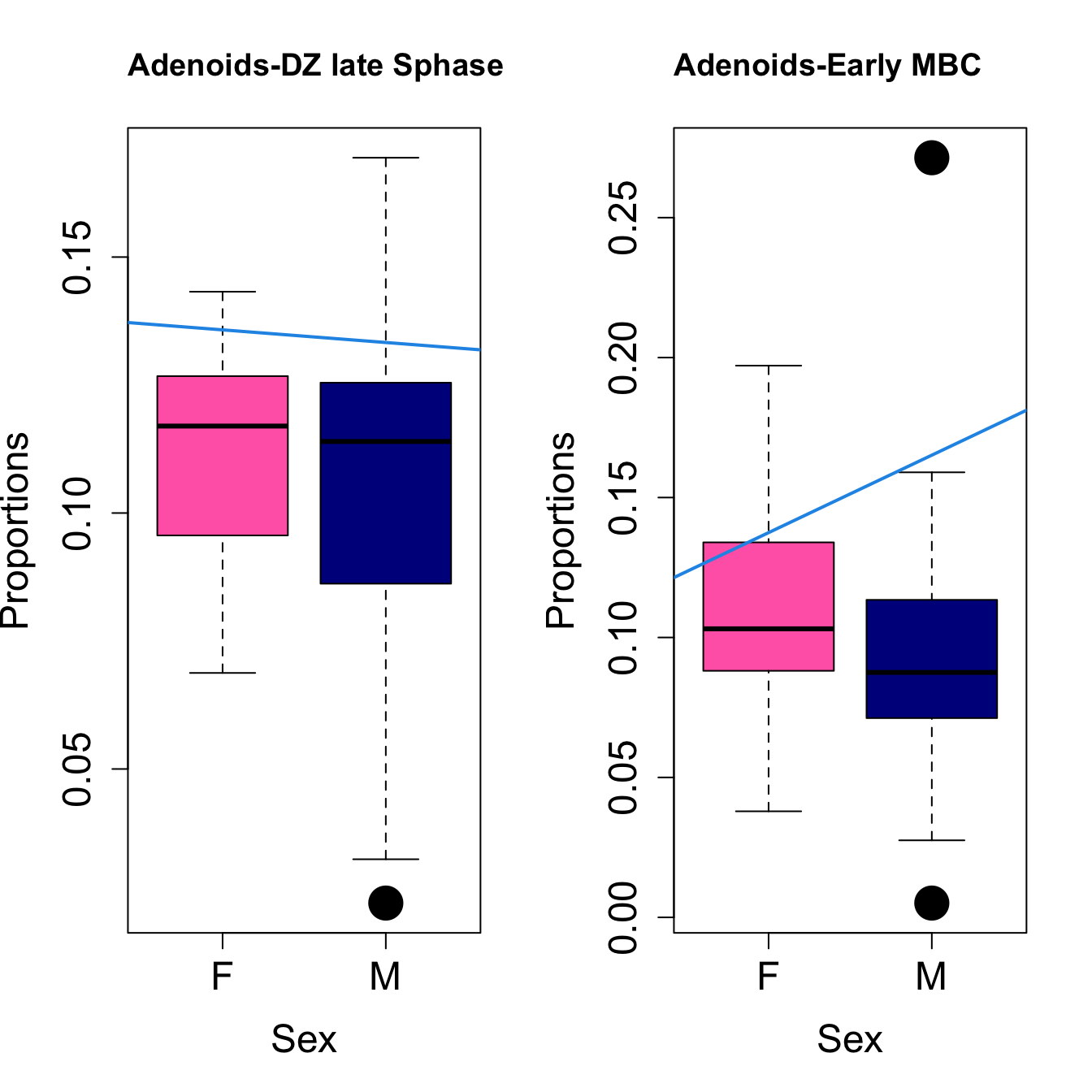
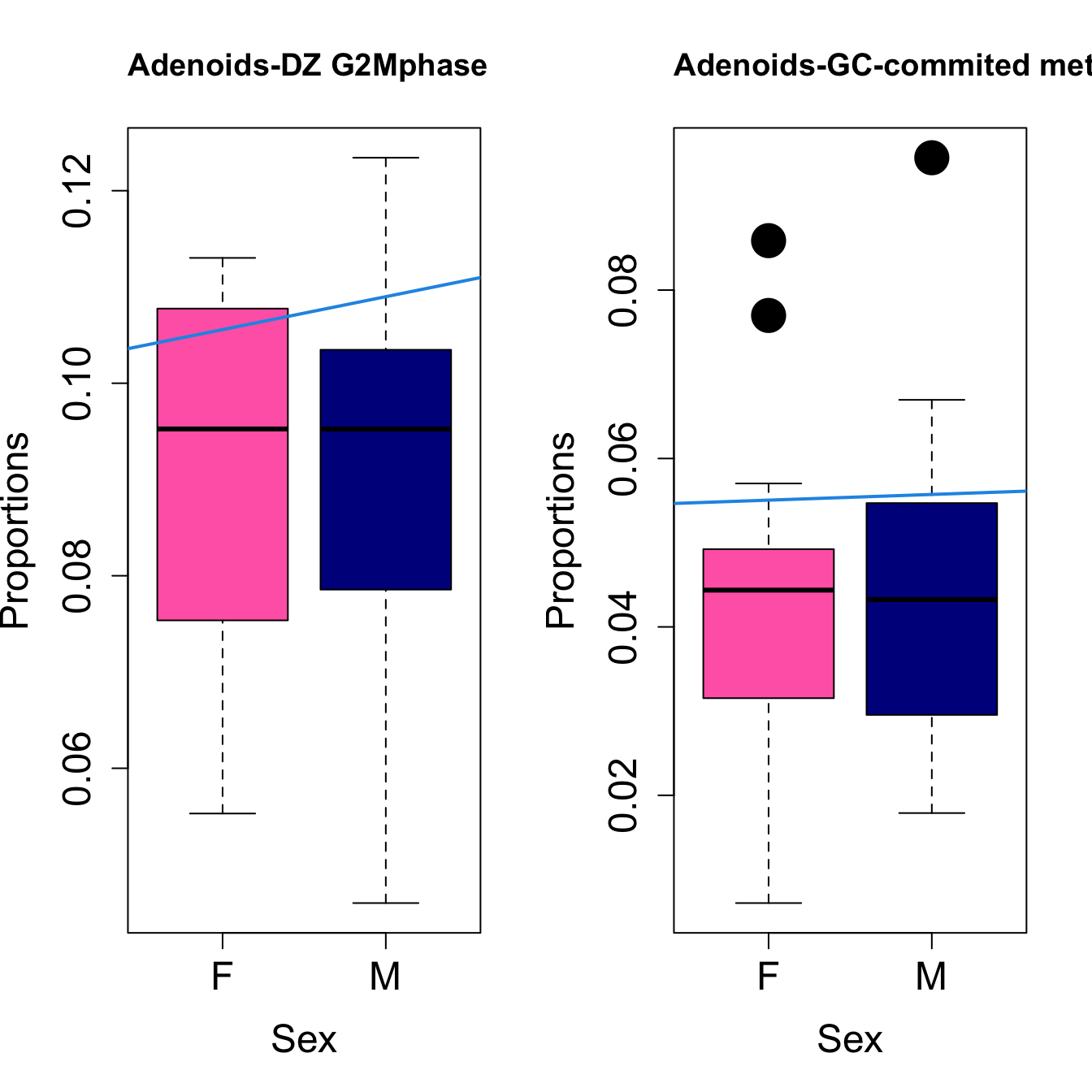
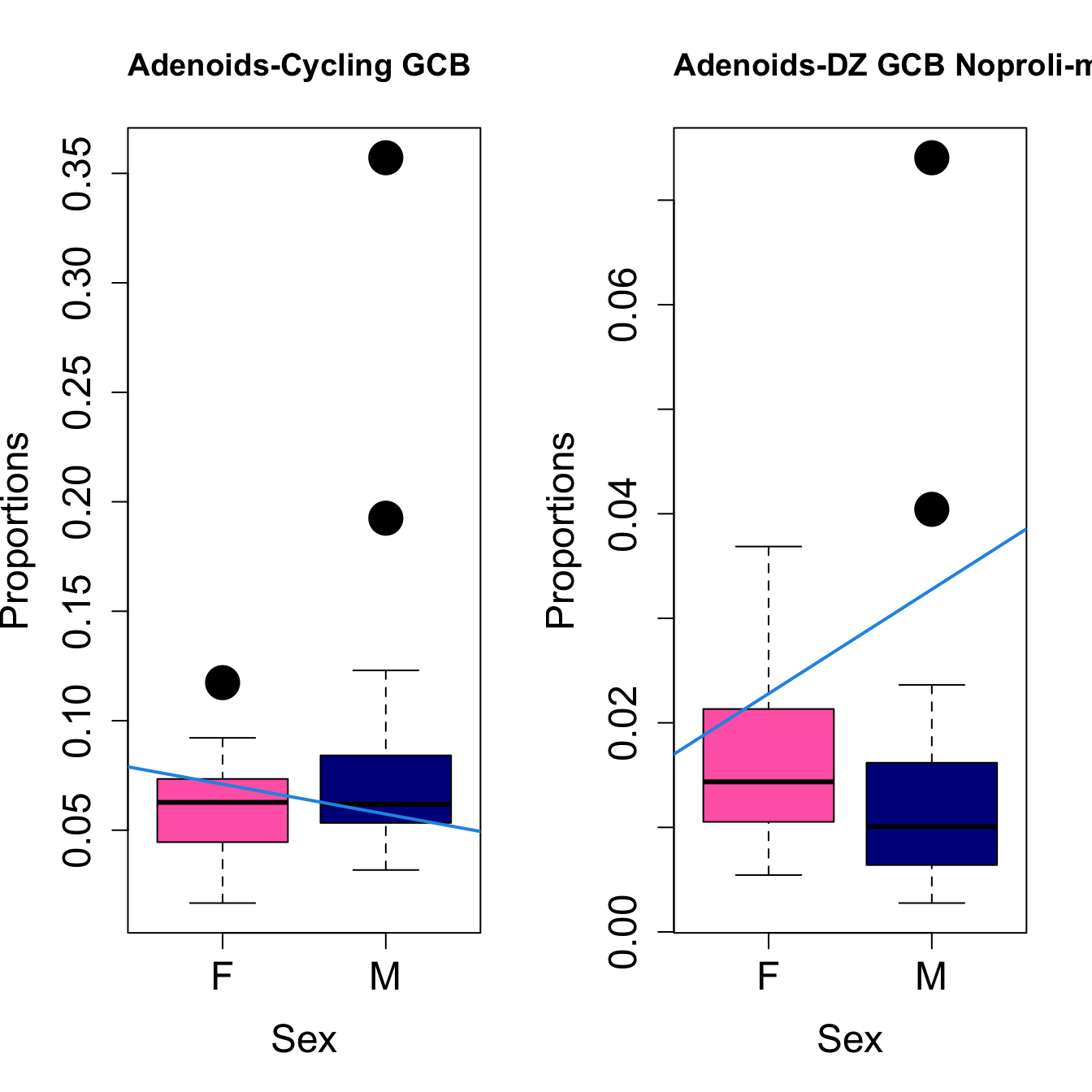
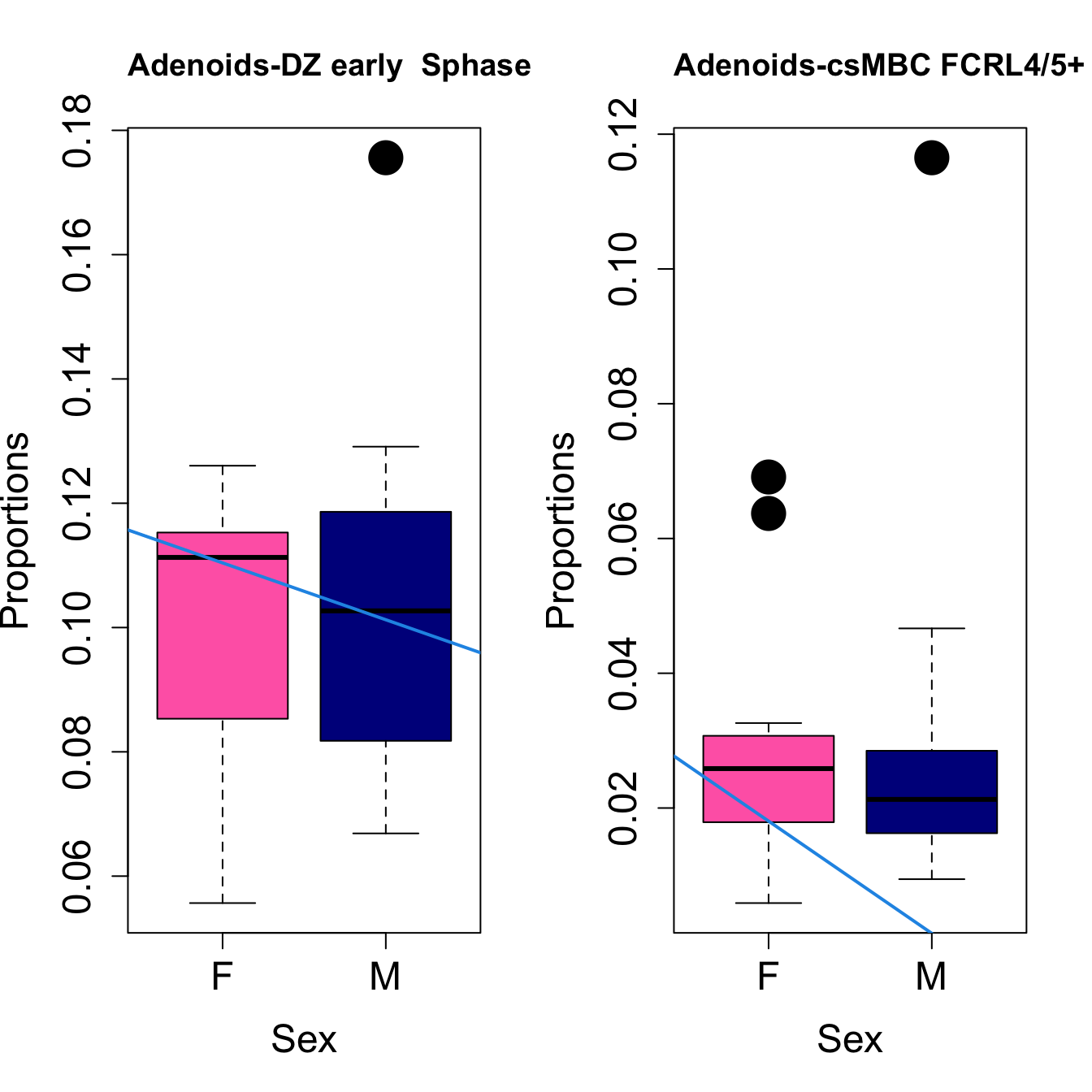
Save results for GCcell population
if (!file.exists(out)) {
write.xlsx(toptable.transformedProps, file = out, rowNames = TRUE, sheetName = "GCcell_subclustering")
} else {
write.xlsx(toptable.transformedProps, file = out, rowNames = TRUE, sheetName = "GCcell_subclustering", append = TRUE)
}Session Info
sessionInfo()R version 4.3.2 (2023-10-31)
Platform: aarch64-apple-darwin20 (64-bit)
Running under: macOS 15.3
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: Australia/Melbourne
tzcode source: internal
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] openxlsx_4.2.5.2 knitr_1.45
[3] kableExtra_1.4.0 edgeR_4.0.16
[5] limma_3.58.1 speckle_1.2.0
[7] ggridges_0.5.6 scran_1.30.2
[9] scuttle_1.12.0 SingleCellExperiment_1.24.0
[11] SummarizedExperiment_1.32.0 Biobase_2.62.0
[13] GenomicRanges_1.54.1 GenomeInfoDb_1.38.6
[15] IRanges_2.36.0 S4Vectors_0.40.2
[17] BiocGenerics_0.48.1 MatrixGenerics_1.14.0
[19] matrixStats_1.2.0 RColorBrewer_1.1-3
[21] ggforce_0.4.2 viridis_0.6.5
[23] viridisLite_0.4.2 paletteer_1.6.0
[25] gridExtra_2.3 lubridate_1.9.3
[27] forcats_1.0.0 stringr_1.5.1
[29] purrr_1.0.2 readr_2.1.5
[31] tidyr_1.3.1 tibble_3.2.1
[33] ggplot2_3.5.0 tidyverse_2.0.0
[35] dplyr_1.1.4 Seurat_5.0.1.9009
[37] SeuratObject_5.0.1 sp_2.1-3
[39] patchwork_1.2.0 glue_1.7.0
[41] here_1.0.1 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] RcppAnnoy_0.0.22 splines_4.3.2
[3] later_1.3.2 bitops_1.0-7
[5] polyclip_1.10-6 fastDummies_1.7.3
[7] lifecycle_1.0.4 rprojroot_2.0.4
[9] globals_0.16.2 processx_3.8.3
[11] lattice_0.22-5 MASS_7.3-60.0.1
[13] magrittr_2.0.3 plotly_4.10.4
[15] sass_0.4.8 rmarkdown_2.25
[17] jquerylib_0.1.4 yaml_2.3.8
[19] metapod_1.10.1 httpuv_1.6.14
[21] sctransform_0.4.1 zip_2.3.1
[23] spam_2.10-0 spatstat.sparse_3.0-3
[25] reticulate_1.35.0 cowplot_1.1.3
[27] pbapply_1.7-2 abind_1.4-5
[29] zlibbioc_1.48.0 Rtsne_0.17
[31] RCurl_1.98-1.14 tweenr_2.0.3
[33] git2r_0.33.0 GenomeInfoDbData_1.2.11
[35] ggrepel_0.9.5 irlba_2.3.5.1
[37] listenv_0.9.1 spatstat.utils_3.0-4
[39] goftest_1.2-3 RSpectra_0.16-1
[41] dqrng_0.3.2 spatstat.random_3.2-2
[43] fitdistrplus_1.1-11 parallelly_1.37.0
[45] svglite_2.1.3 DelayedMatrixStats_1.24.0
[47] DelayedArray_0.28.0 leiden_0.4.3.1
[49] codetools_0.2-19 xml2_1.3.6
[51] tidyselect_1.2.0 farver_2.1.1
[53] ScaledMatrix_1.10.0 spatstat.explore_3.2-6
[55] jsonlite_1.8.8 BiocNeighbors_1.20.2
[57] ellipsis_0.3.2 progressr_0.14.0
[59] survival_3.5-8 systemfonts_1.0.5
[61] tools_4.3.2 ica_1.0-3
[63] Rcpp_1.0.12 SparseArray_1.2.4
[65] xfun_0.42 withr_3.0.0
[67] fastmap_1.1.1 bluster_1.12.0
[69] fansi_1.0.6 rsvd_1.0.5
[71] callr_3.7.5 digest_0.6.34
[73] timechange_0.3.0 R6_2.5.1
[75] mime_0.12 colorspace_2.1-0
[77] scattermore_1.2 tensor_1.5
[79] spatstat.data_3.0-4 utf8_1.2.4
[81] generics_0.1.3 data.table_1.15.0
[83] httr_1.4.7 htmlwidgets_1.6.4
[85] S4Arrays_1.2.0 whisker_0.4.1
[87] uwot_0.1.16 pkgconfig_2.0.3
[89] gtable_0.3.4 lmtest_0.9-40
[91] XVector_0.42.0 htmltools_0.5.7
[93] dotCall64_1.1-1 scales_1.3.0
[95] png_0.1-8 rstudioapi_0.15.0
[97] tzdb_0.4.0 reshape2_1.4.4
[99] nlme_3.1-164 cachem_1.0.8
[101] zoo_1.8-12 KernSmooth_2.23-22
[103] parallel_4.3.2 miniUI_0.1.1.1
[105] pillar_1.9.0 grid_4.3.2
[107] vctrs_0.6.5 RANN_2.6.1
[109] promises_1.2.1 BiocSingular_1.18.0
[111] beachmat_2.18.1 xtable_1.8-4
[113] cluster_2.1.6 evaluate_0.23
[115] locfit_1.5-9.8 cli_3.6.2
[117] compiler_4.3.2 rlang_1.1.3
[119] crayon_1.5.2 future.apply_1.11.1
[121] labeling_0.4.3 rematch2_2.1.2
[123] ps_1.7.6 getPass_0.2-4
[125] plyr_1.8.9 fs_1.6.3
[127] stringi_1.8.3 BiocParallel_1.36.0
[129] deldir_2.0-2 munsell_0.5.0
[131] lazyeval_0.2.2 spatstat.geom_3.2-8
[133] Matrix_1.6-5 RcppHNSW_0.6.0
[135] hms_1.1.3 sparseMatrixStats_1.14.0
[137] future_1.33.1 statmod_1.5.0
[139] shiny_1.8.0 highr_0.10
[141] ROCR_1.0-11 igraph_2.0.2
[143] bslib_0.6.1
sessionInfo()R version 4.3.2 (2023-10-31)
Platform: aarch64-apple-darwin20 (64-bit)
Running under: macOS 15.3
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: Australia/Melbourne
tzcode source: internal
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] openxlsx_4.2.5.2 knitr_1.45
[3] kableExtra_1.4.0 edgeR_4.0.16
[5] limma_3.58.1 speckle_1.2.0
[7] ggridges_0.5.6 scran_1.30.2
[9] scuttle_1.12.0 SingleCellExperiment_1.24.0
[11] SummarizedExperiment_1.32.0 Biobase_2.62.0
[13] GenomicRanges_1.54.1 GenomeInfoDb_1.38.6
[15] IRanges_2.36.0 S4Vectors_0.40.2
[17] BiocGenerics_0.48.1 MatrixGenerics_1.14.0
[19] matrixStats_1.2.0 RColorBrewer_1.1-3
[21] ggforce_0.4.2 viridis_0.6.5
[23] viridisLite_0.4.2 paletteer_1.6.0
[25] gridExtra_2.3 lubridate_1.9.3
[27] forcats_1.0.0 stringr_1.5.1
[29] purrr_1.0.2 readr_2.1.5
[31] tidyr_1.3.1 tibble_3.2.1
[33] ggplot2_3.5.0 tidyverse_2.0.0
[35] dplyr_1.1.4 Seurat_5.0.1.9009
[37] SeuratObject_5.0.1 sp_2.1-3
[39] patchwork_1.2.0 glue_1.7.0
[41] here_1.0.1 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] RcppAnnoy_0.0.22 splines_4.3.2
[3] later_1.3.2 bitops_1.0-7
[5] polyclip_1.10-6 fastDummies_1.7.3
[7] lifecycle_1.0.4 rprojroot_2.0.4
[9] globals_0.16.2 processx_3.8.3
[11] lattice_0.22-5 MASS_7.3-60.0.1
[13] magrittr_2.0.3 plotly_4.10.4
[15] sass_0.4.8 rmarkdown_2.25
[17] jquerylib_0.1.4 yaml_2.3.8
[19] metapod_1.10.1 httpuv_1.6.14
[21] sctransform_0.4.1 zip_2.3.1
[23] spam_2.10-0 spatstat.sparse_3.0-3
[25] reticulate_1.35.0 cowplot_1.1.3
[27] pbapply_1.7-2 abind_1.4-5
[29] zlibbioc_1.48.0 Rtsne_0.17
[31] RCurl_1.98-1.14 tweenr_2.0.3
[33] git2r_0.33.0 GenomeInfoDbData_1.2.11
[35] ggrepel_0.9.5 irlba_2.3.5.1
[37] listenv_0.9.1 spatstat.utils_3.0-4
[39] goftest_1.2-3 RSpectra_0.16-1
[41] dqrng_0.3.2 spatstat.random_3.2-2
[43] fitdistrplus_1.1-11 parallelly_1.37.0
[45] svglite_2.1.3 DelayedMatrixStats_1.24.0
[47] DelayedArray_0.28.0 leiden_0.4.3.1
[49] codetools_0.2-19 xml2_1.3.6
[51] tidyselect_1.2.0 farver_2.1.1
[53] ScaledMatrix_1.10.0 spatstat.explore_3.2-6
[55] jsonlite_1.8.8 BiocNeighbors_1.20.2
[57] ellipsis_0.3.2 progressr_0.14.0
[59] survival_3.5-8 systemfonts_1.0.5
[61] tools_4.3.2 ica_1.0-3
[63] Rcpp_1.0.12 SparseArray_1.2.4
[65] xfun_0.42 withr_3.0.0
[67] fastmap_1.1.1 bluster_1.12.0
[69] fansi_1.0.6 rsvd_1.0.5
[71] callr_3.7.5 digest_0.6.34
[73] timechange_0.3.0 R6_2.5.1
[75] mime_0.12 colorspace_2.1-0
[77] scattermore_1.2 tensor_1.5
[79] spatstat.data_3.0-4 utf8_1.2.4
[81] generics_0.1.3 data.table_1.15.0
[83] httr_1.4.7 htmlwidgets_1.6.4
[85] S4Arrays_1.2.0 whisker_0.4.1
[87] uwot_0.1.16 pkgconfig_2.0.3
[89] gtable_0.3.4 lmtest_0.9-40
[91] XVector_0.42.0 htmltools_0.5.7
[93] dotCall64_1.1-1 scales_1.3.0
[95] png_0.1-8 rstudioapi_0.15.0
[97] tzdb_0.4.0 reshape2_1.4.4
[99] nlme_3.1-164 cachem_1.0.8
[101] zoo_1.8-12 KernSmooth_2.23-22
[103] parallel_4.3.2 miniUI_0.1.1.1
[105] pillar_1.9.0 grid_4.3.2
[107] vctrs_0.6.5 RANN_2.6.1
[109] promises_1.2.1 BiocSingular_1.18.0
[111] beachmat_2.18.1 xtable_1.8-4
[113] cluster_2.1.6 evaluate_0.23
[115] locfit_1.5-9.8 cli_3.6.2
[117] compiler_4.3.2 rlang_1.1.3
[119] crayon_1.5.2 future.apply_1.11.1
[121] labeling_0.4.3 rematch2_2.1.2
[123] ps_1.7.6 getPass_0.2-4
[125] plyr_1.8.9 fs_1.6.3
[127] stringi_1.8.3 BiocParallel_1.36.0
[129] deldir_2.0-2 munsell_0.5.0
[131] lazyeval_0.2.2 spatstat.geom_3.2-8
[133] Matrix_1.6-5 RcppHNSW_0.6.0
[135] hms_1.1.3 sparseMatrixStats_1.14.0
[137] future_1.33.1 statmod_1.5.0
[139] shiny_1.8.0 highr_0.10
[141] ROCR_1.0-11 igraph_2.0.2
[143] bslib_0.6.1