Paedriatic airway atlas: all Tissues
Quality control: Exploratory Plots
Gunjan Dixit and Jovana Maksimovic
May 23, 2024
Last updated: 2024-05-23
Checks: 6 1
Knit directory: paed-airway-allTissues/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R
Markdown file created these results, you’ll want to first commit it to
the Git repo. If you’re still working on the analysis, you can ignore
this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230811) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 5aee5dd. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: data/.DS_Store
Ignored: data/RDS/
Ignored: output/.DS_Store
Ignored: output/CSV/.DS_Store
Ignored: output/G000231_Neeland_batch1/
Ignored: output/G000231_Neeland_batch2_1/
Ignored: output/G000231_Neeland_batch2_2/
Ignored: output/G000231_Neeland_batch3/
Ignored: output/G000231_Neeland_batch4/
Ignored: output/G000231_Neeland_batch5/
Ignored: output/G000231_Neeland_batch9_1/
Ignored: output/RDS/
Ignored: output/plots/
Untracked files:
Untracked: analysis/03_Batch_Integration.Rmd
Untracked: analysis/Age_modeling.Rmd
Untracked: analysis/Age_proportions.Rmd
Untracked: analysis/Age_proportions_AllBatches.Rmd
Untracked: analysis/AllBatches_QCExploratory.Rmd
Untracked: analysis/Batch_Integration_&_Downstream_analysis.Rmd
Untracked: analysis/Batch_correction_&_Downstream.Rmd
Untracked: analysis/Cell_cycle_regression.Rmd
Untracked: analysis/Preprocessing_Batch1_Nasal_brushings.Rmd
Untracked: analysis/Preprocessing_Batch2_Tonsils.Rmd
Untracked: analysis/Preprocessing_Batch3_Adenoids.Rmd
Untracked: analysis/Preprocessing_Batch4_Bronchial_brushings.Rmd
Untracked: analysis/Preprocessing_Batch5_Nasal_brushings.Rmd
Untracked: analysis/Preprocessing_Batch6_BAL.Rmd
Untracked: analysis/Preprocessing_Batch7_Bronchial_brushings.Rmd
Untracked: analysis/Preprocessing_Batch8_Adenoids.Rmd
Untracked: analysis/Preprocessing_Batch9_Tonsils.Rmd
Untracked: analysis/cell_cycle_regression.R
Untracked: analysis/test.Rmd
Untracked: analysis/testing_age_all.Rmd
Untracked: data/Hs.c2.cp.reactome.v7.1.entrez.rds
Untracked: data/Raw_feature_bc_matrix/
Untracked: data/celltypes_Mel_GD_v3.xlsx
Untracked: data/celltypes_Mel_GD_v4_no_dups.xlsx
Untracked: data/celltypes_Mel_modified.xlsx
Untracked: data/celltypes_Mel_v2.csv
Untracked: data/celltypes_Mel_v2.xlsx
Untracked: data/celltypes_Mel_v2_MN.xlsx
Untracked: data/celltypes_for_mel_MN.xlsx
Untracked: data/earlyAIR_sample_sheets_combined.xlsx
Untracked: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/
Untracked: output/CSV/Bronchial_brushings_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/
Untracked: output/CSV/Nasal_brushings_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/
Unstaged changes:
Deleted: 02_QC_exploratoryPlots.Rmd
Deleted: 02_QC_exploratoryPlots.html
Modified: analysis/00_AllBatches_overview.Rmd
Modified: analysis/01_QC_emptyDrops.Rmd
Modified: analysis/BAL.Rmd
Modified: analysis/Bronchial_brushings.Rmd
Modified: analysis/Nasal_brushings.Rmd
Modified: analysis/index.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with
wflow_publish() to start tracking its development.
Introduction
This RMarkdown contains exploratory plots to determine the quality of scRNA-seq data obtained from healthy tissue samples from five key locations of the respiratory system in children. The aim of this project is to develop a cell atlas of the healthy pediatric airway.
Load libraries
suppressPackageStartupMessages({
library(BiocStyle)
library(tidyverse)
library(here)
library(glue)
library(RColorBrewer)
library(scran)
library(scater)
library(Seurat)
library(scuttle)
library(janitor)
library(cowplot)
library(patchwork)
library(scales)
library(Homo.sapiens)
library(msigdbr)
library(EnsDb.Hsapiens.v86)
library(ensembldb)
})Load data
List the batches.
Batch 1- Nasal brushings (n=16)
Batch
2- Tonsils (n=16)
Batch 3- Adenoids
(n=16)
Batch 4- Bronchial brushings (n=16)
Batch 5- Nasal brushings (n=16)
Batch
6- BAL (n=8*2)
Batch 7- Bronchial
brushings (n=16)
Batch 8- Adenoids (n=16)
Batch 9- Tonsils (n=16)
batches_beforeQC <- dir(here("output/RDS/AllBatches_filtered_SCEs/"), full.names = TRUE, pattern = "_filtered.SCE.rds")
batches_afterQC <- dir(here("output/RDS/AllBatches_Azimuth_SEUs/"), full.names = TRUE, pattern = "filter.Azimuth.SEU.rds")
batches <- data.frame(Before = batches_beforeQC, After = batches_afterQC)Non-empty droplets from CellRanger
Read CellRanger filtered output for each batch and summarize them together using bar plot.
plot_cellnum <- function(sample_data, title) {
num_unique_samples <- length(unique(sample_data$Sample))
color_palette <- colorRampPalette(brewer.pal(min(9, num_unique_samples), "Set3"))(num_unique_samples)
names(color_palette) <- sort(unique(sample_data$Sample))
p <- ggplot(sample_data, aes(x = Sample, fill = Sample)) +
geom_bar(stat = "count") +
coord_flip() +
labs(y = "Number of droplets", title = title) +
theme_cowplot(font_size = 10) +
geom_text(stat = 'count', aes(label = ..count..), hjust = 1.5, size = 3) +
guides(fill = guide_legend(title = "Sample")) +
scale_fill_manual(values = color_palette)
return(p)
}
plot_list <- list()
for (i in seq_along(batches$Before)) {
sce <- readRDS(batches$Before[i])
sce1 <- as.SingleCellExperiment(readRDS(batches$After[i]))
batch_name <- gsub(pattern = ".*/|\\.CellRanger.*", replacement = "", x = batches$Before[i])
p1 <- plot_cellnum(data.frame(Sample = sce$Sample), paste0(batch_name, " Before QC"))
p2 <- plot_cellnum(data.frame(Sample = sce1$Sample), paste0(batch_name, " After QC"))
plot_list[[batch_name]] <- plot_grid(p1, p2, nrow = 2)
}
# Print the plots within markdown tabs
for (batch_name in names(plot_list)) {
cat("###", batch_name, "\n")
print(plot_list[[batch_name]])
cat("\n")
}G000231_batch1_Nasal_brushings
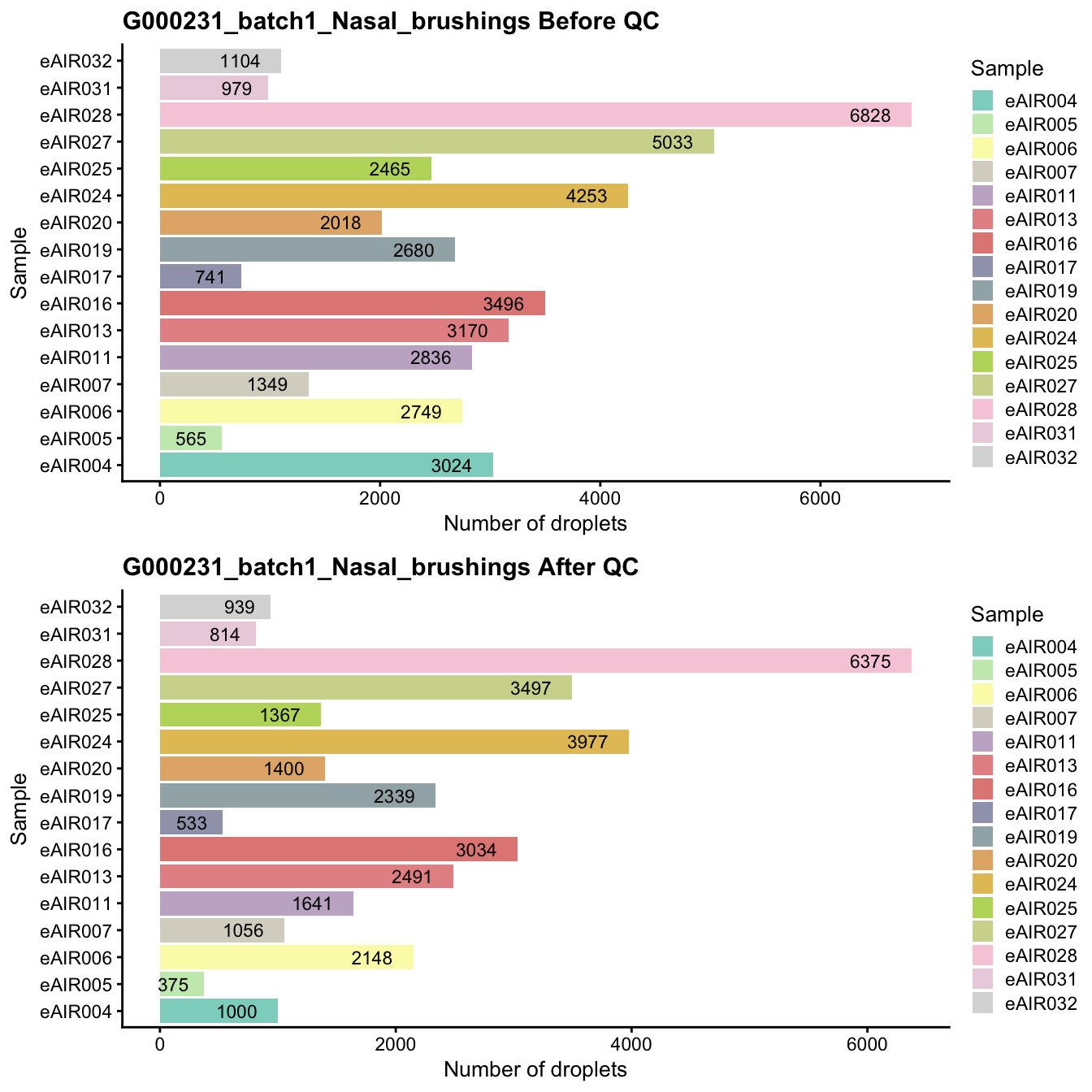
Breakdown of the samples.
G000231_batch2_Tonsils
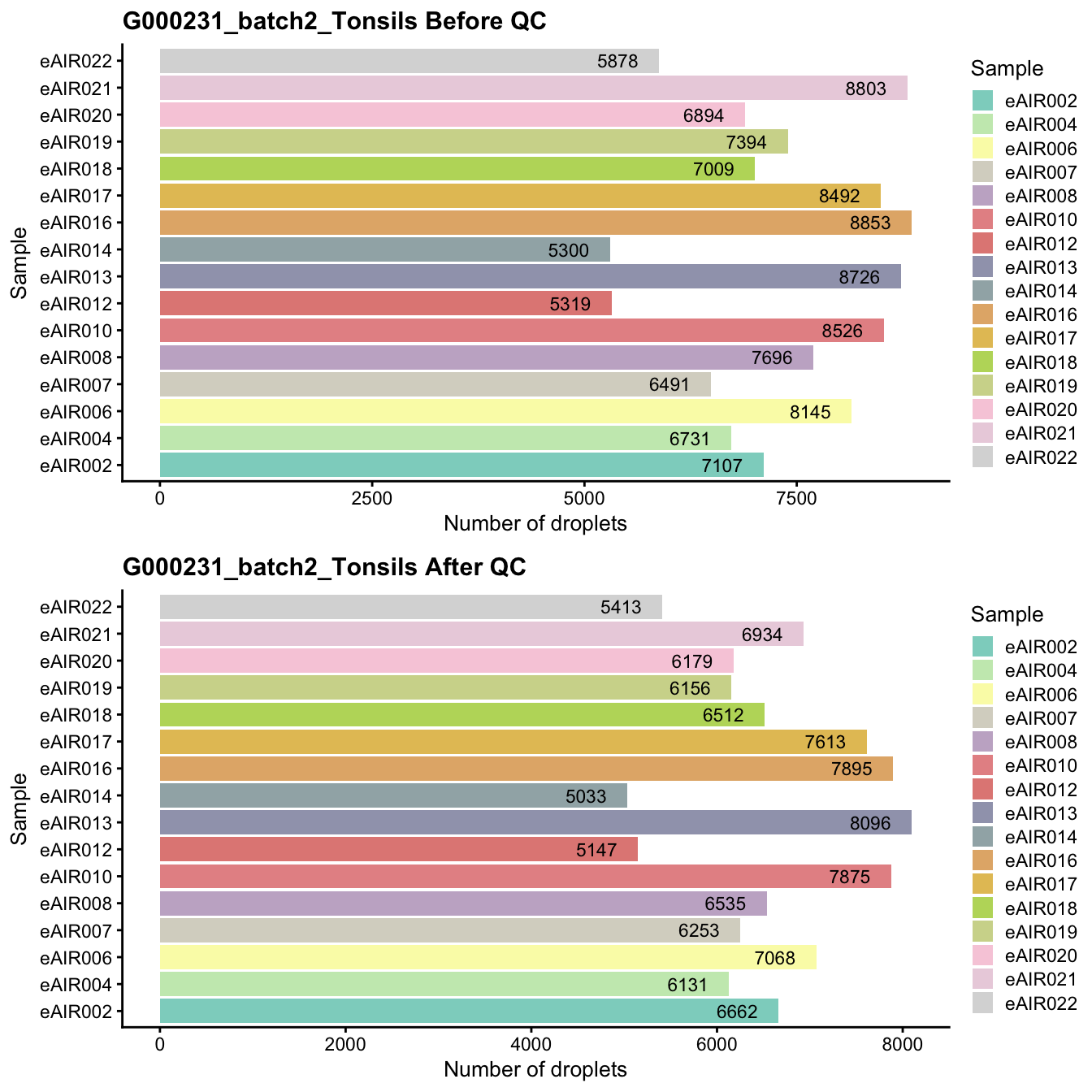
Breakdown of the samples.
G000231_batch3_Adenoids
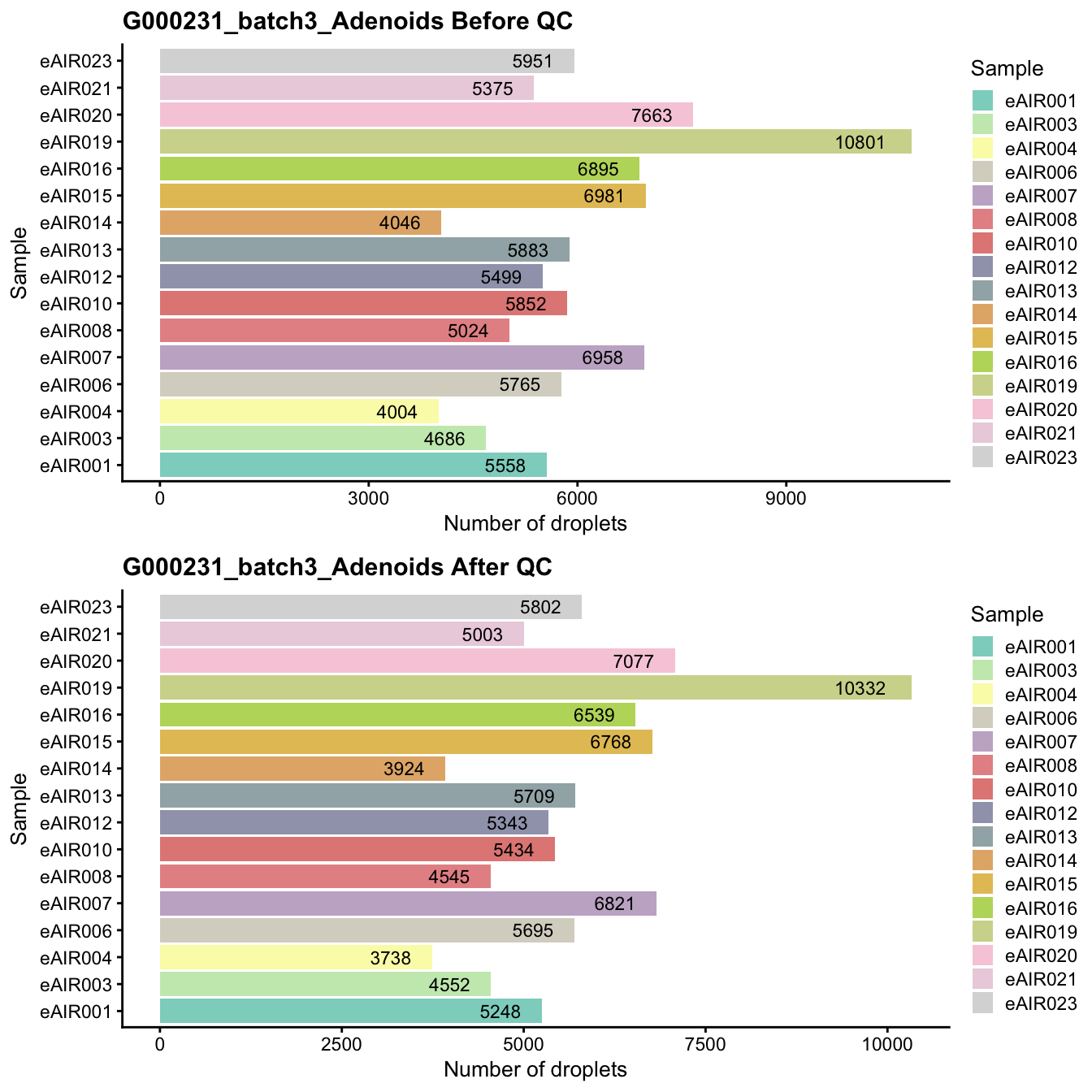
Breakdown of the samples.
G000231_batch4_Bronchial_brushings
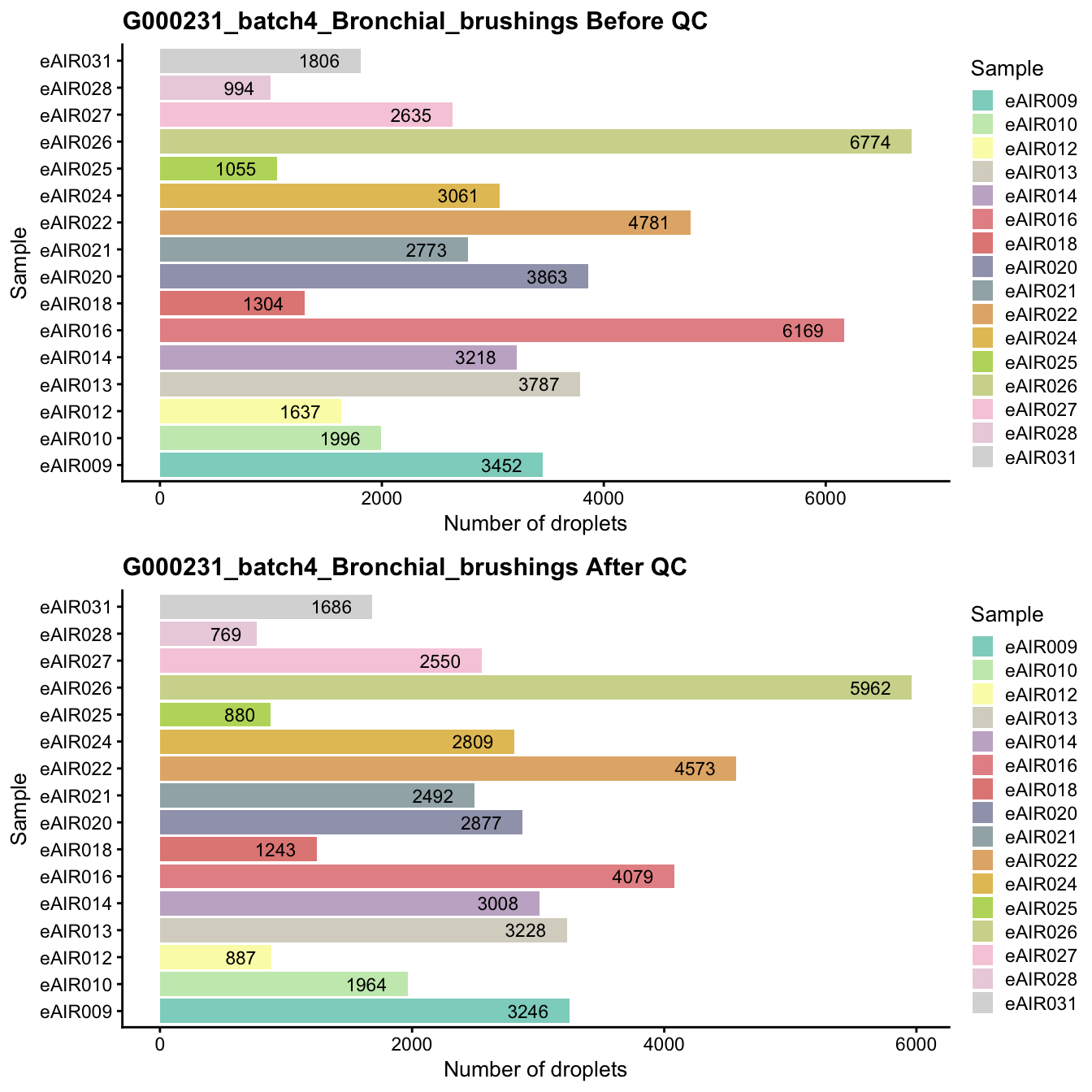
Breakdown of the samples.
G000231_batch5_Nasal_brushings
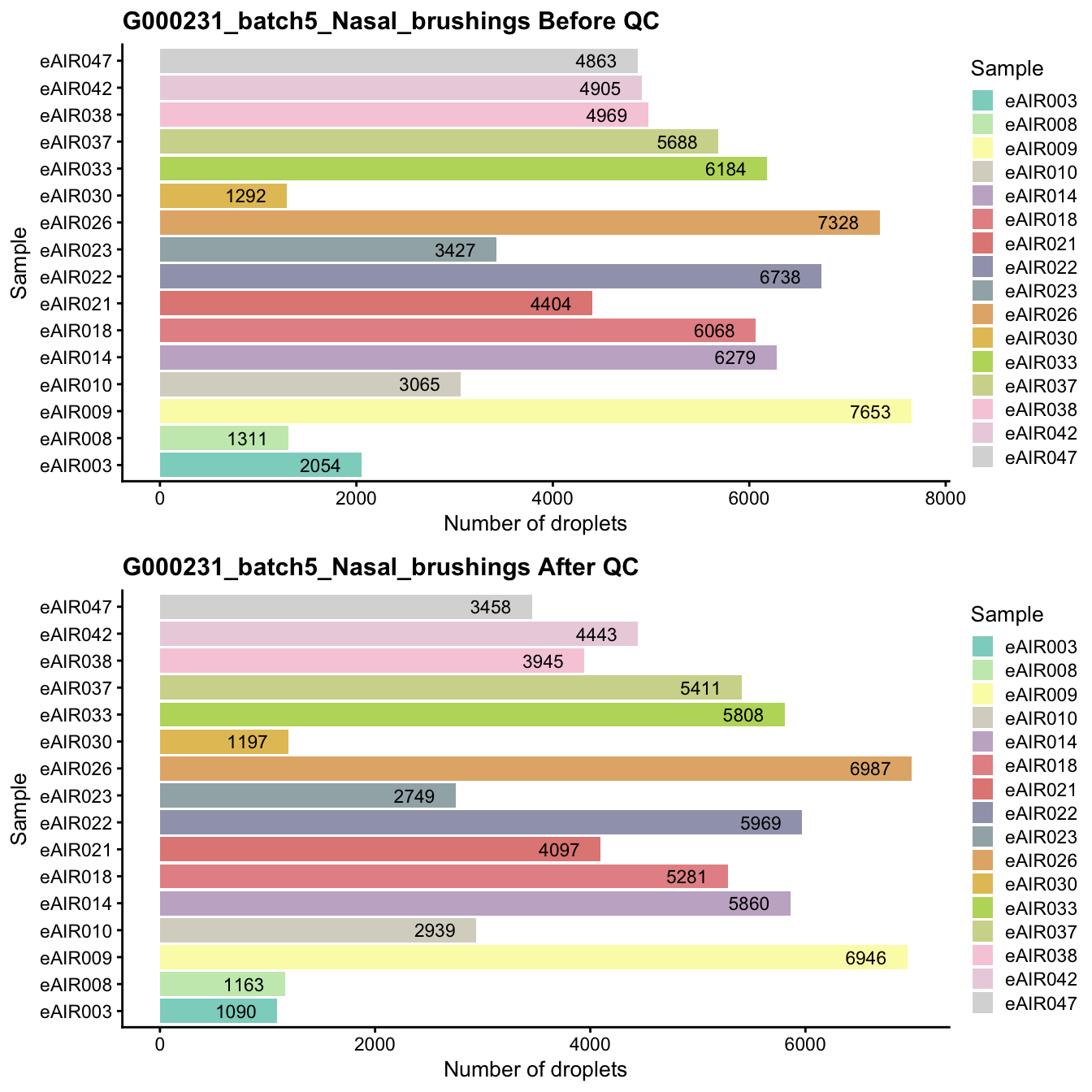
Breakdown of the samples.
G000231_batch6_BAL
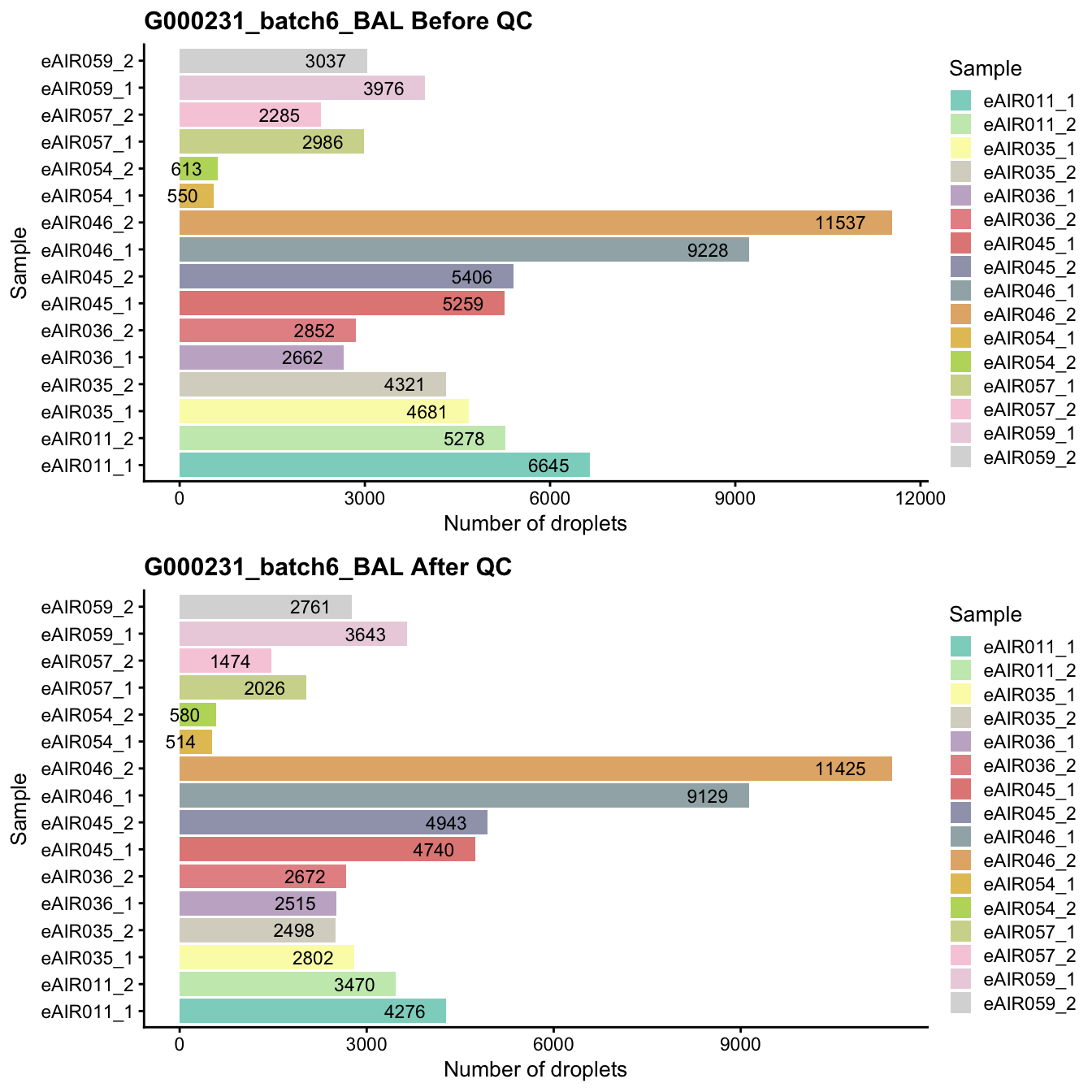
Breakdown of the samples.
G000231_batch7_Bronchial_brushings
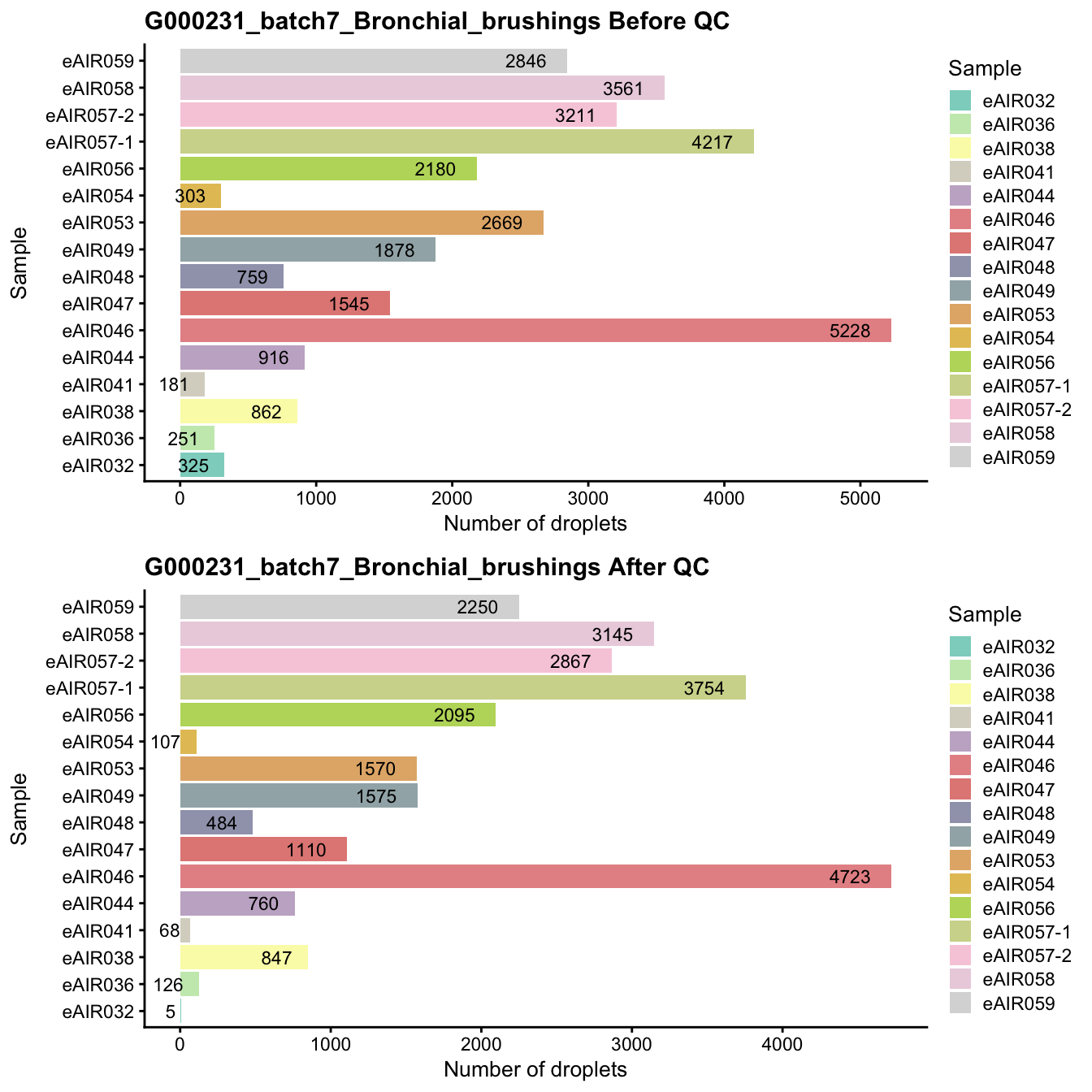
Breakdown of the samples.
G000231_batch8_Adenoids
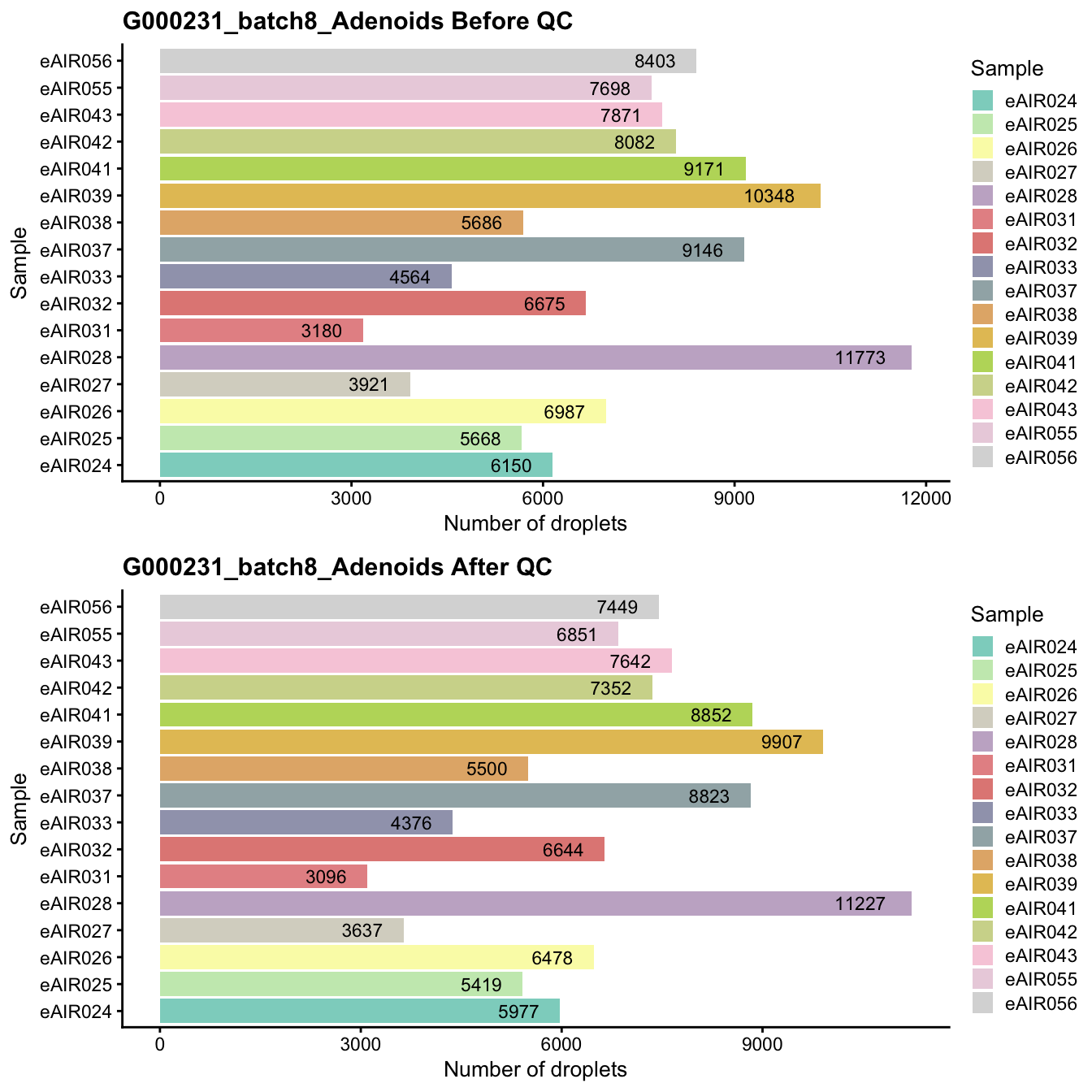
Breakdown of the samples.
G000231_batch9_Tonsils
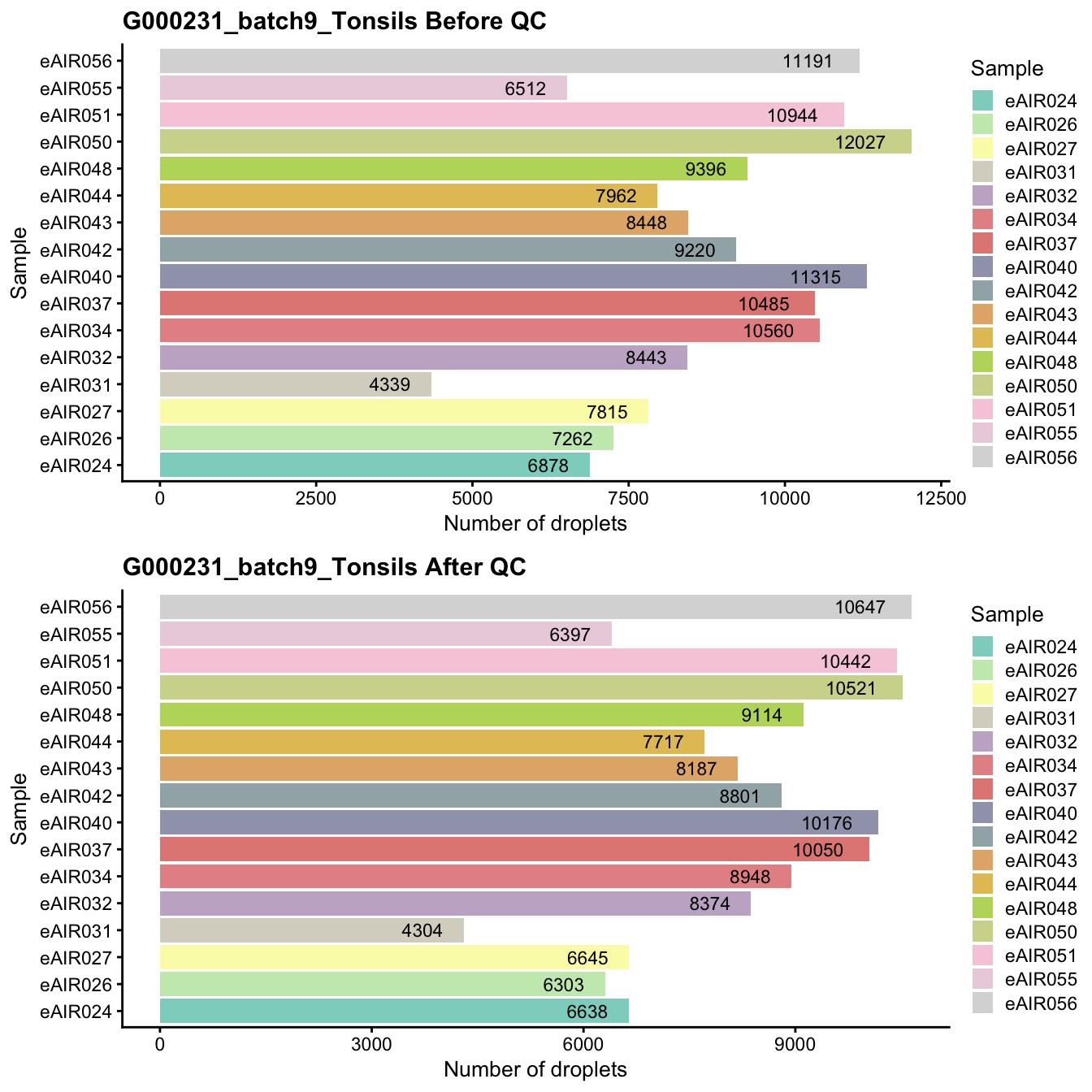
Breakdown of the samples.
Library Size and detected genes before and after applying QC measures-
plot_qc_metrics <- function(sce, title) {
q1 <- plotColData(
sce,
"sum",
x = "Sample",
colour_by = "Sample",
point_size = 0.5) +
scale_y_log10() +
annotation_logticks(
sides = "l",
short = unit(0.03, "cm"),
mid = unit(0.06, "cm"),
long = unit(0.09, "cm")) +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0))
q2 <- plotColData(
sce,
"detected",
x = "Sample",
colour_by = "Sample",
point_size = 0.5) +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0))
qc_plots <- (q1 + q2) + plot_layout(guides = "collect", ncol = 2) +
theme(legend.position = "none")
return(qc_plots)
}
plot_list <- list()
for (i in seq_along(batches$Before)) {
sce_before <- readRDS(batches$Before[i])
is.mito <- grepl(pattern = "^MT", rownames(sce_before))
sce_before <- addPerCellQCMetrics(sce_before, subsets = list(mito = is.mito))
sce_after <- as.SingleCellExperiment(readRDS(batches$After[i]))
is.mito <- grepl(pattern = "^MT", rownames(sce_after))
sce_after <- addPerCellQCMetrics(sce_after, subsets = list(mito = is.mito))
batch_name <- gsub(pattern = ".*/|\\.CellRanger.*", replacement = "", x = batches$Before[i])
# Generating QC plots and calculating medians
q1 <- plot_qc_metrics(sce_before, paste0(batch_name, " Before QC"))
q2 <- plot_qc_metrics(sce_after, paste0(batch_name, " After QC"))
plot_list[[batch_name]] <- plot_grid(q1, q2, nrow = 2)
}
# Print the plots within markdown tabs
for (batch_name in names(plot_list)) {
cat("###", batch_name, "\n")
print(batch_name)
print(plot_list[[batch_name]])
cat("\n")
}G000231_batch1_Nasal_brushings
[1] “G000231_batch1_Nasal_brushings”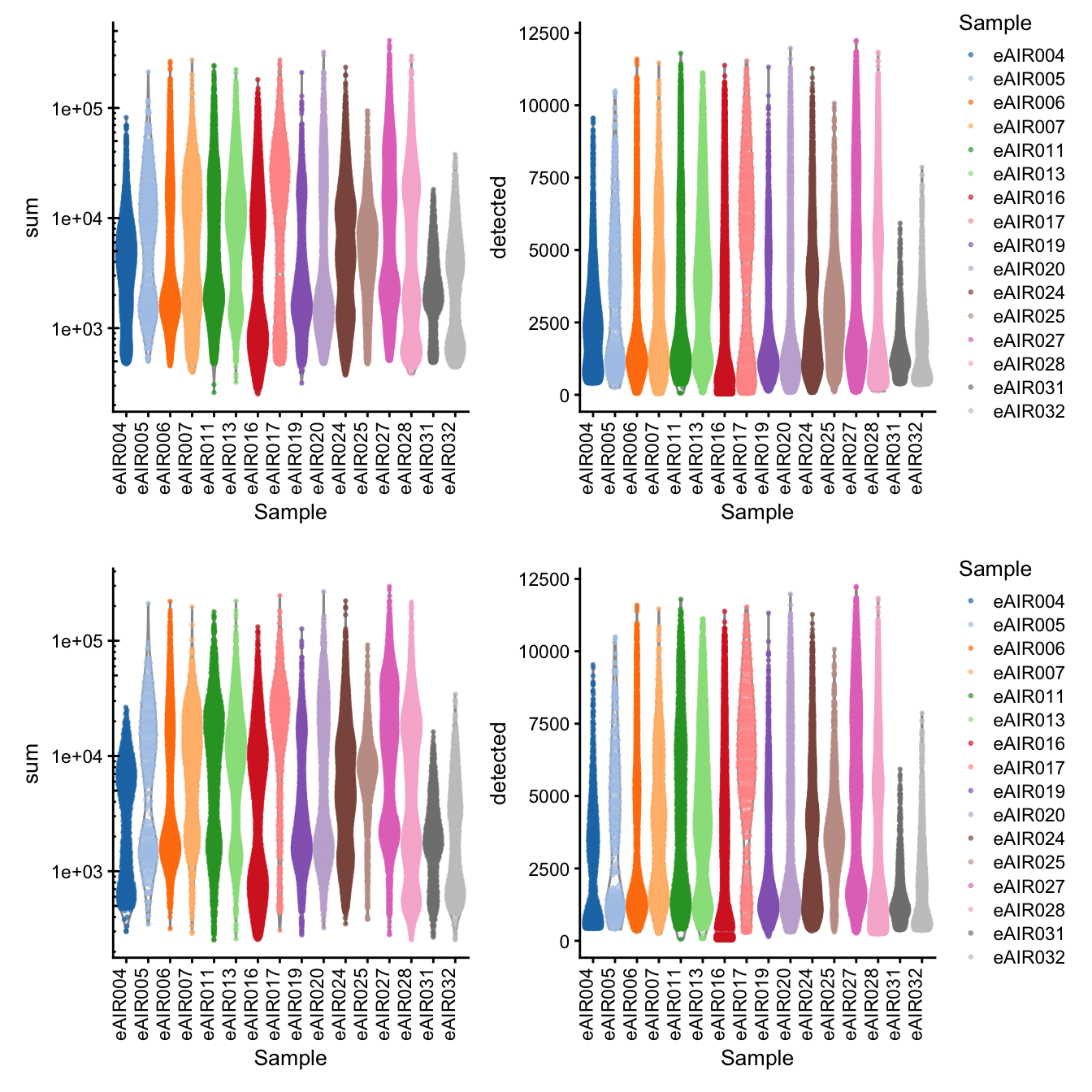
Library Size and detected genes of the samples before and after QC.
G000231_batch2_Tonsils
[1] “G000231_batch2_Tonsils”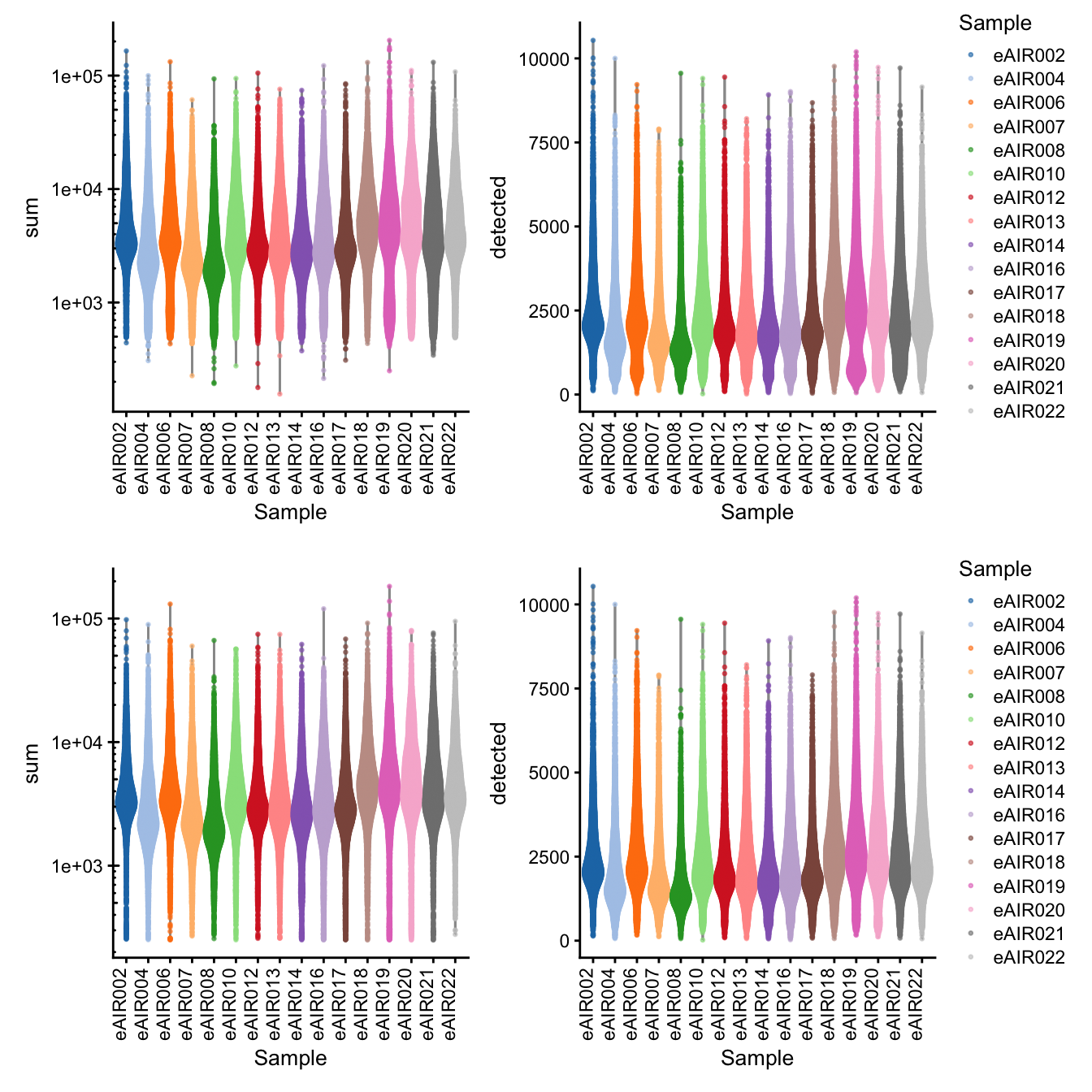
Library Size and detected genes of the samples before and after QC.
G000231_batch3_Adenoids
[1] “G000231_batch3_Adenoids”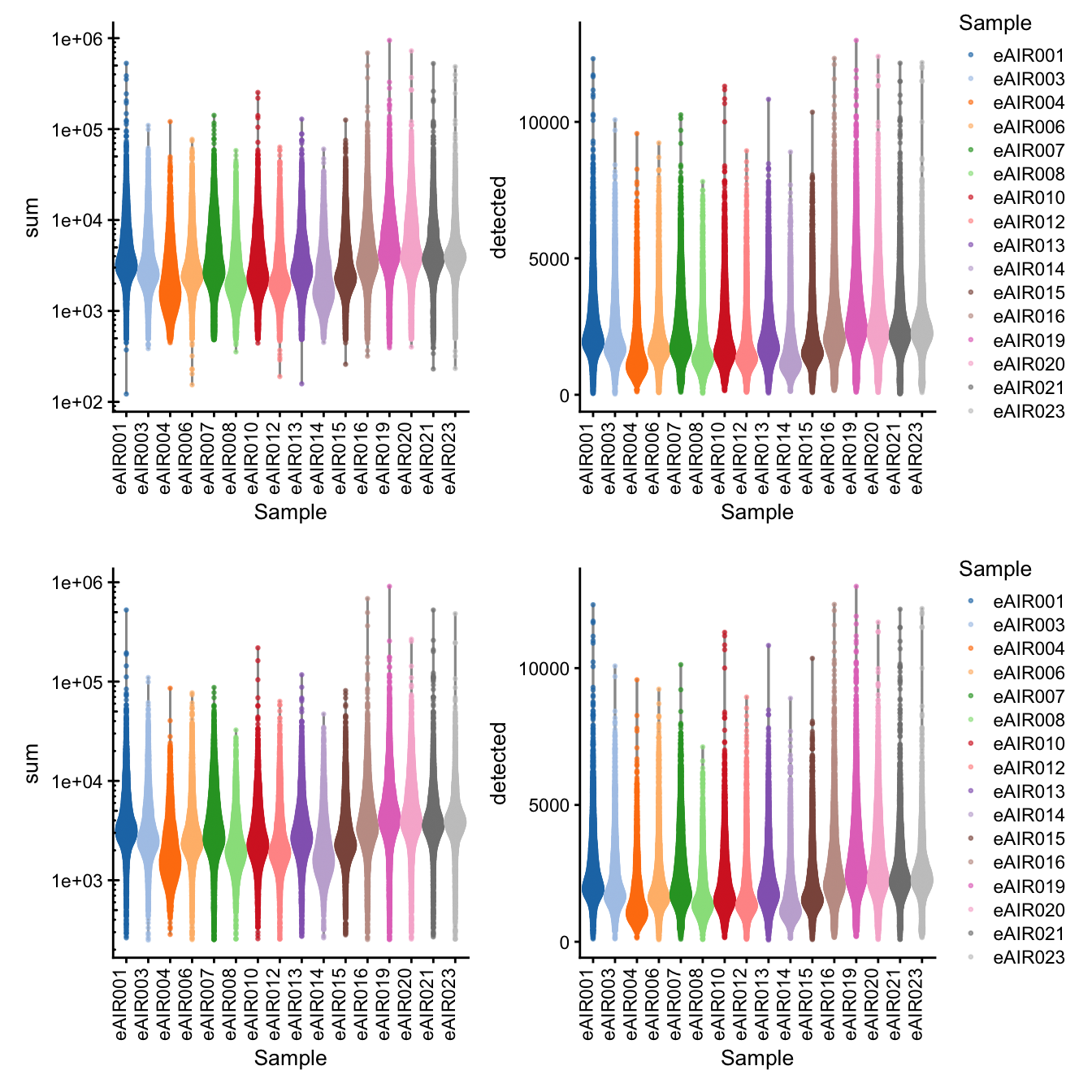
Library Size and detected genes of the samples before and after QC.
G000231_batch4_Bronchial_brushings
[1] “G000231_batch4_Bronchial_brushings”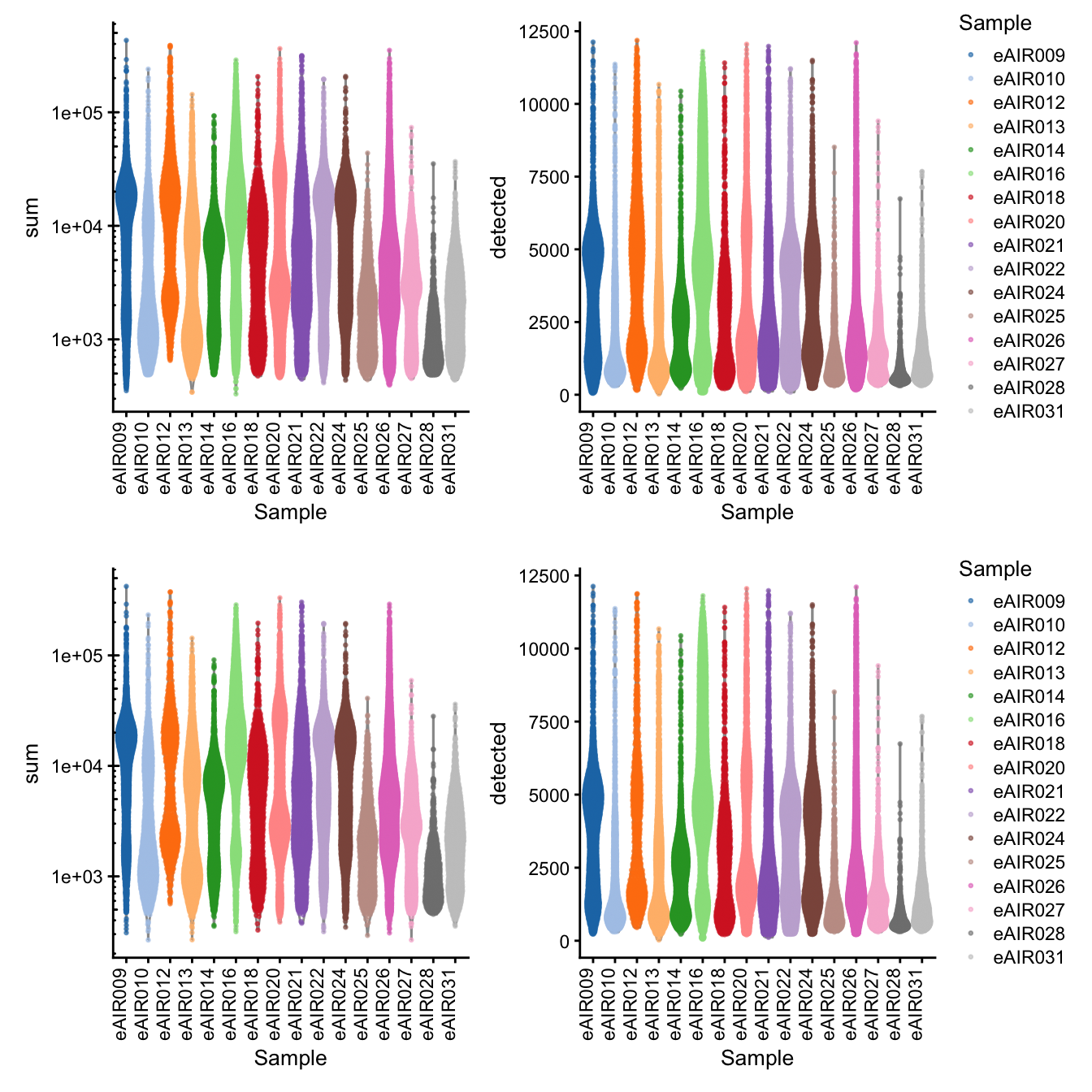
Library Size and detected genes of the samples before and after QC.
G000231_batch5_Nasal_brushings
[1] “G000231_batch5_Nasal_brushings”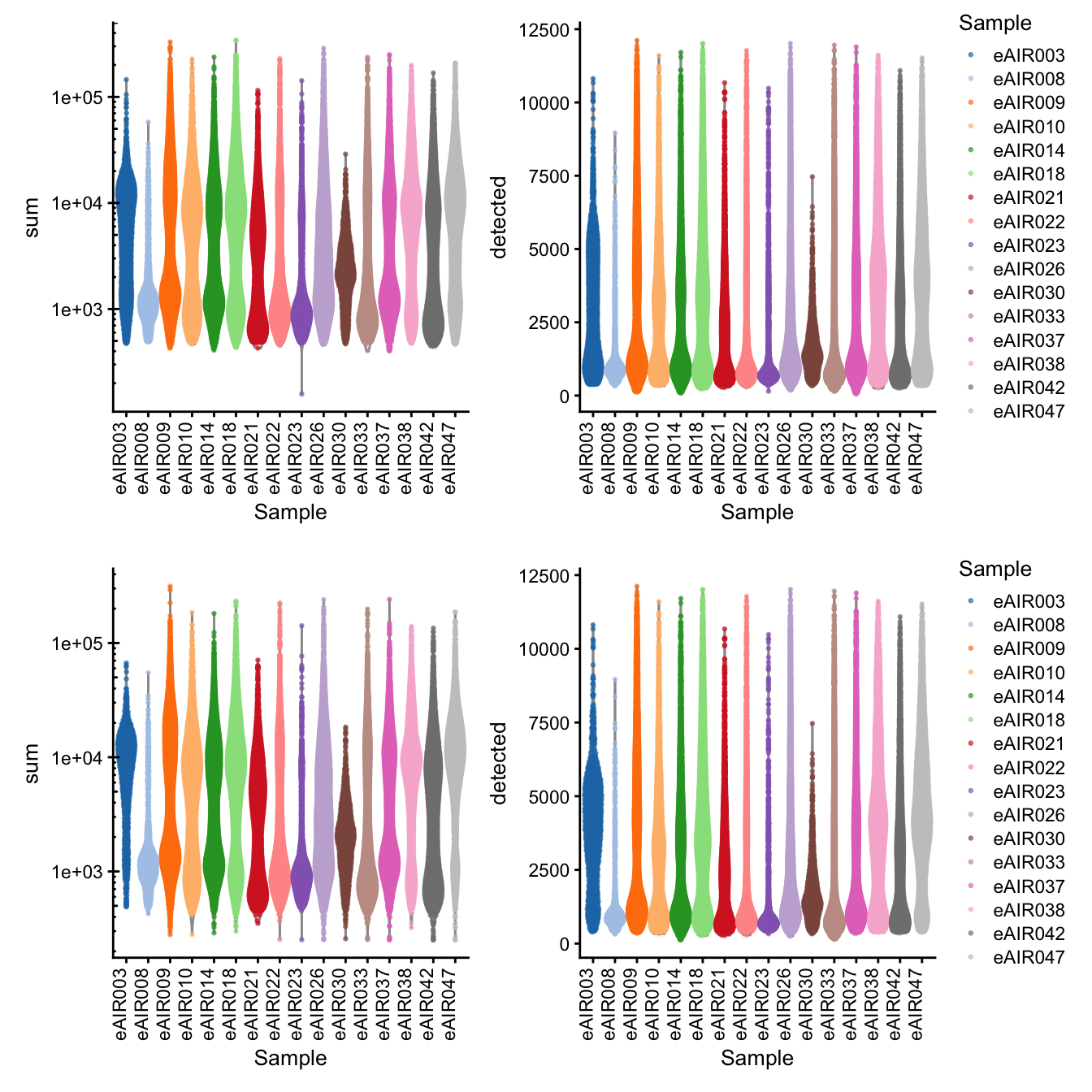
Library Size and detected genes of the samples before and after QC.
G000231_batch6_BAL
[1] “G000231_batch6_BAL”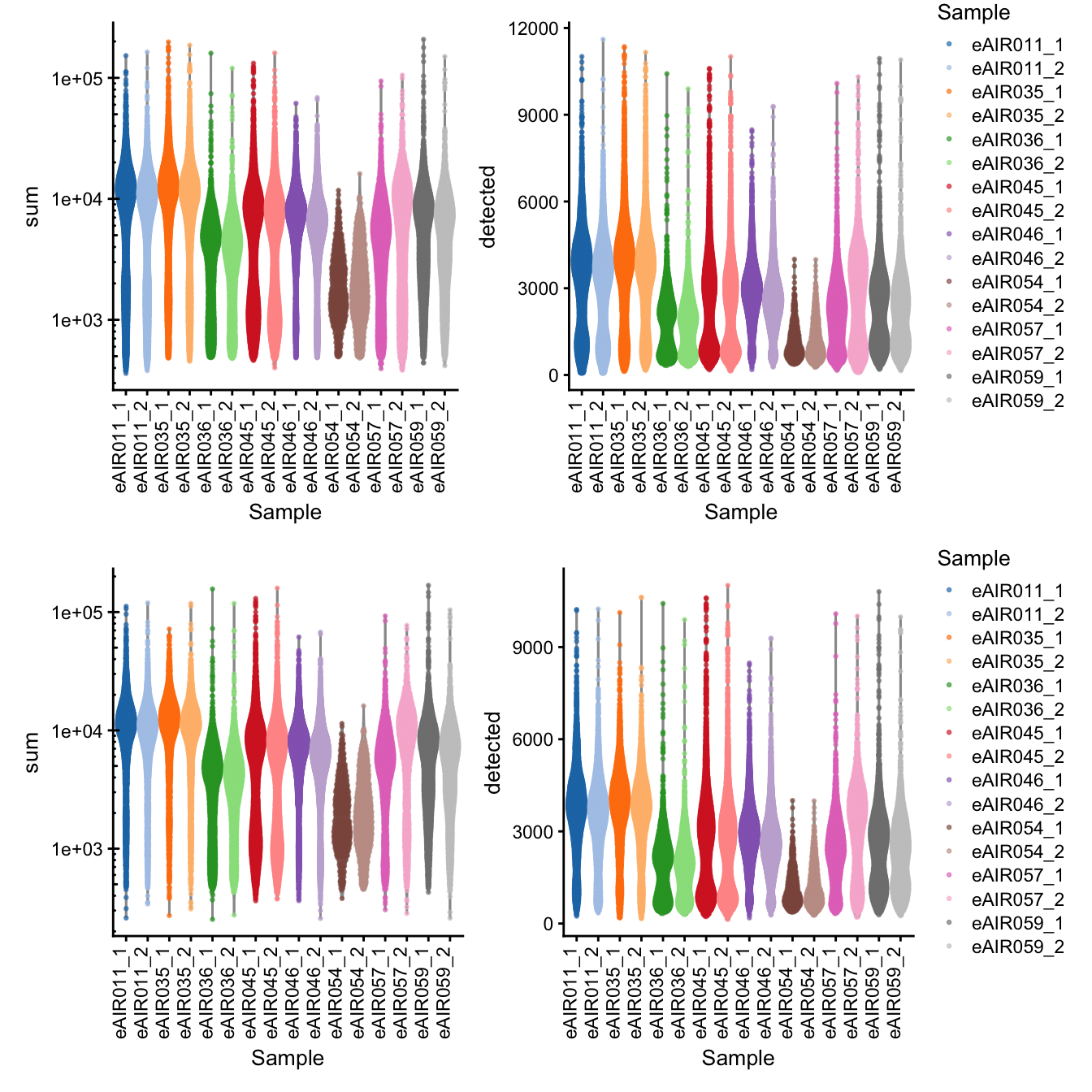
Library Size and detected genes of the samples before and after QC.
G000231_batch7_Bronchial_brushings
[1] “G000231_batch7_Bronchial_brushings”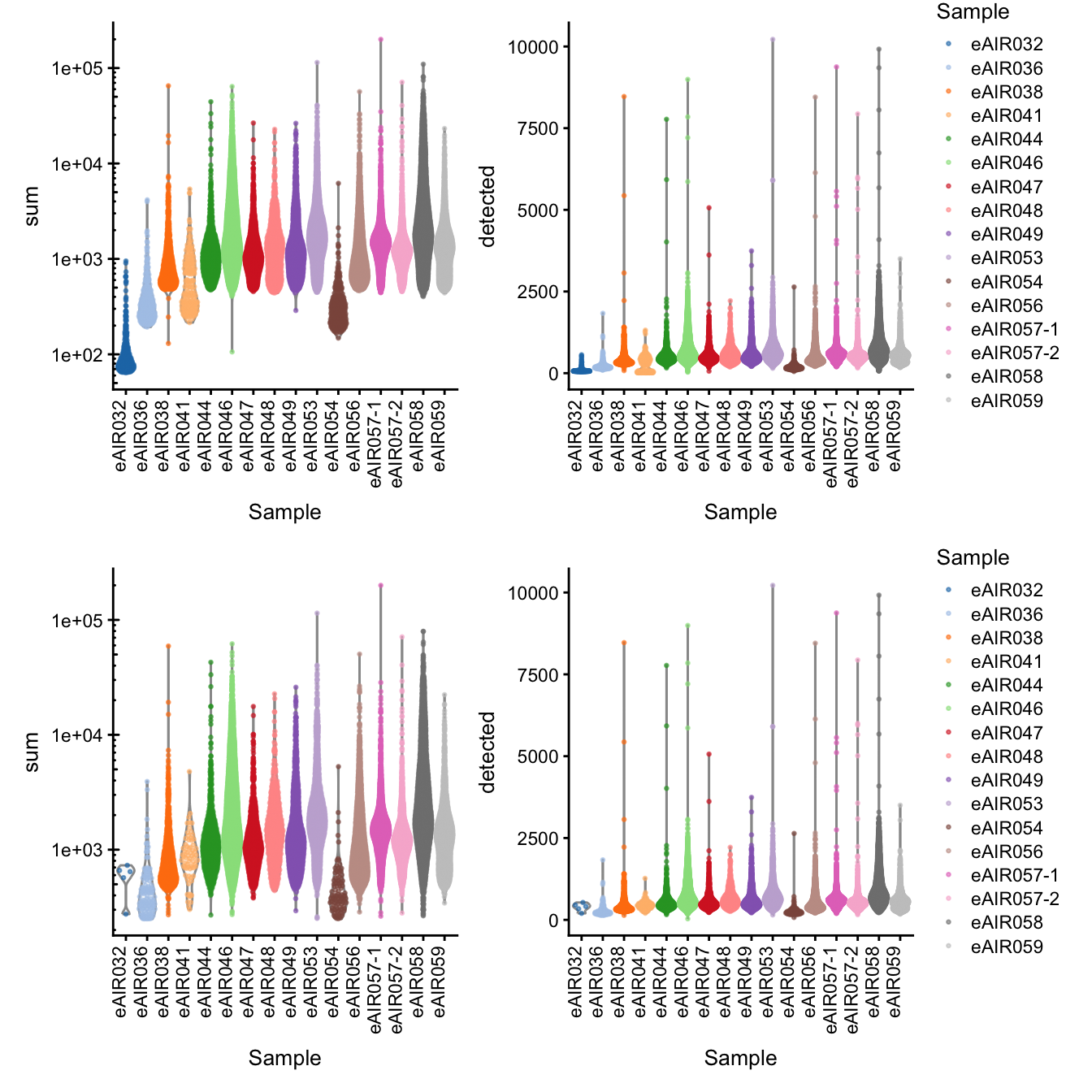
Library Size and detected genes of the samples before and after QC.
G000231_batch8_Adenoids
[1] “G000231_batch8_Adenoids”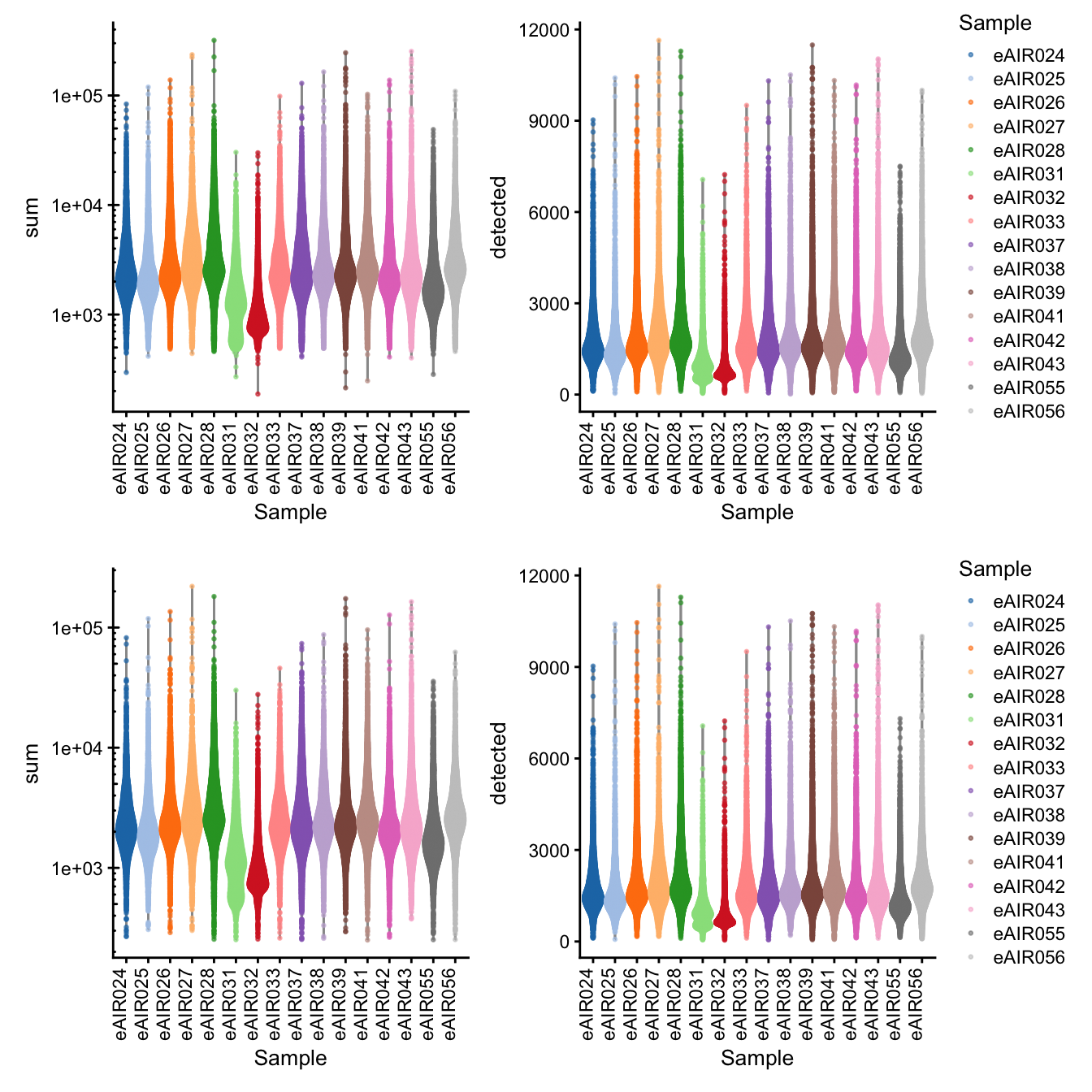
Library Size and detected genes of the samples before and after QC.
G000231_batch9_Tonsils
[1] “G000231_batch9_Tonsils”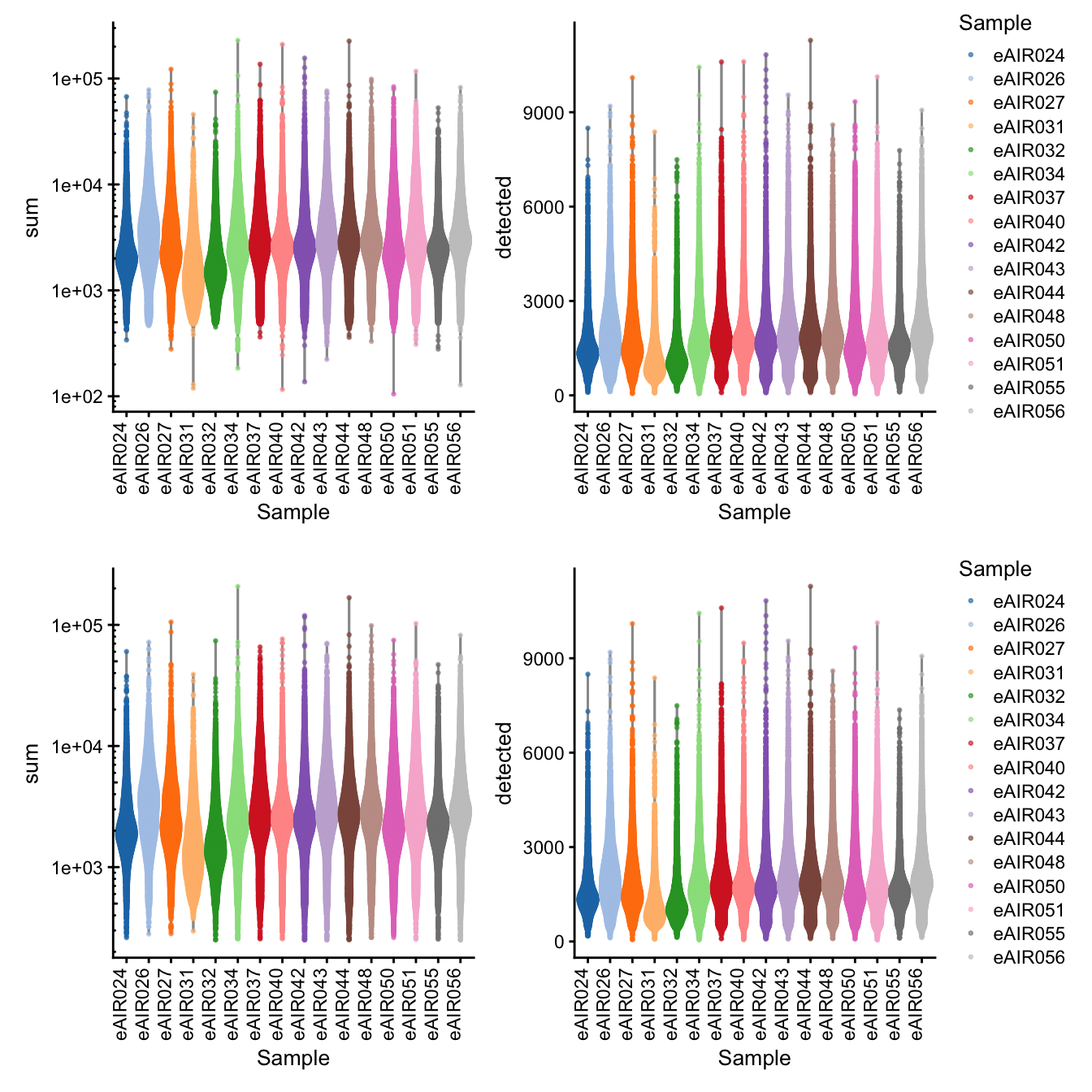
Library Size and detected genes of the samples before and after QC.
Session Info
sessioninfo::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.3.2 (2023-10-31)
os macOS Sonoma 14.5
system aarch64, darwin20
ui X11
language (EN)
collate en_US.UTF-8
ctype en_US.UTF-8
tz Australia/Melbourne
date 2024-05-23
pandoc 3.1.1 @ /Users/dixitgunjan/Desktop/RStudio.app/Contents/Resources/app/quarto/bin/tools/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
abind 1.4-5 2016-07-21 [1] CRAN (R 4.3.0)
AnnotationDbi * 1.64.1 2023-11-02 [1] Bioconductor
AnnotationFilter * 1.26.0 2023-10-26 [1] Bioconductor
babelgene 22.9 2022-09-29 [1] CRAN (R 4.3.0)
beachmat 2.18.1 2024-02-17 [1] Bioconductor 3.18 (R 4.3.2)
beeswarm 0.4.0 2021-06-01 [1] CRAN (R 4.3.0)
Biobase * 2.62.0 2023-10-26 [1] Bioconductor
BiocFileCache 2.10.1 2023-10-26 [1] Bioconductor
BiocGenerics * 0.48.1 2023-11-02 [1] Bioconductor
BiocIO 1.12.0 2023-10-26 [1] Bioconductor
BiocManager 1.30.22 2023-08-08 [1] CRAN (R 4.3.0)
BiocNeighbors 1.20.2 2024-01-13 [1] Bioconductor 3.18 (R 4.3.2)
BiocParallel 1.36.0 2023-10-26 [1] Bioconductor
BiocSingular 1.18.0 2023-11-06 [1] Bioconductor
BiocStyle * 2.30.0 2023-10-26 [1] Bioconductor
biomaRt 2.58.2 2024-02-03 [1] Bioconductor 3.18 (R 4.3.2)
Biostrings 2.70.2 2024-01-30 [1] Bioconductor 3.18 (R 4.3.2)
bit 4.0.5 2022-11-15 [1] CRAN (R 4.3.0)
bit64 4.0.5 2020-08-30 [1] CRAN (R 4.3.0)
bitops 1.0-7 2021-04-24 [1] CRAN (R 4.3.0)
blob 1.2.4 2023-03-17 [1] CRAN (R 4.3.0)
bluster 1.12.0 2023-12-19 [1] Bioconductor 3.18 (R 4.3.2)
bslib 0.6.1 2023-11-28 [1] CRAN (R 4.3.1)
cachem 1.0.8 2023-05-01 [1] CRAN (R 4.3.0)
callr 3.7.5 2024-02-19 [1] CRAN (R 4.3.1)
cli 3.6.2 2023-12-11 [1] CRAN (R 4.3.1)
cluster 2.1.6 2023-12-01 [1] CRAN (R 4.3.1)
codetools 0.2-19 2023-02-01 [1] CRAN (R 4.3.2)
colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.3.0)
cowplot * 1.1.3 2024-01-22 [1] CRAN (R 4.3.1)
crayon 1.5.2 2022-09-29 [1] CRAN (R 4.3.0)
curl 5.2.0 2023-12-08 [1] CRAN (R 4.3.1)
data.table 1.15.0 2024-01-30 [1] CRAN (R 4.3.1)
DBI 1.2.2 2024-02-16 [1] CRAN (R 4.3.1)
dbplyr 2.4.0 2023-10-26 [1] CRAN (R 4.3.1)
DelayedArray 0.28.0 2023-11-06 [1] Bioconductor
DelayedMatrixStats 1.24.0 2023-11-06 [1] Bioconductor
deldir 2.0-2 2023-11-23 [1] CRAN (R 4.3.1)
digest 0.6.34 2024-01-11 [1] CRAN (R 4.3.1)
dotCall64 1.1-1 2023-11-28 [1] CRAN (R 4.3.1)
dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.3.1)
dqrng 0.3.2 2023-11-29 [1] CRAN (R 4.3.1)
edgeR 4.0.16 2024-02-20 [1] Bioconductor 3.18 (R 4.3.2)
ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.3.0)
EnsDb.Hsapiens.v86 * 2.99.0 2024-02-27 [1] Bioconductor
ensembldb * 2.26.0 2023-10-26 [1] Bioconductor
evaluate 0.23 2023-11-01 [1] CRAN (R 4.3.1)
fansi 1.0.6 2023-12-08 [1] CRAN (R 4.3.1)
farver 2.1.1 2022-07-06 [1] CRAN (R 4.3.0)
fastDummies 1.7.3 2023-07-06 [1] CRAN (R 4.3.0)
fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.3.0)
filelock 1.0.3 2023-12-11 [1] CRAN (R 4.3.1)
fitdistrplus 1.1-11 2023-04-25 [1] CRAN (R 4.3.0)
forcats * 1.0.0 2023-01-29 [1] CRAN (R 4.3.0)
fs 1.6.3 2023-07-20 [1] CRAN (R 4.3.0)
future 1.33.1 2023-12-22 [1] CRAN (R 4.3.1)
future.apply 1.11.1 2023-12-21 [1] CRAN (R 4.3.1)
generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
GenomeInfoDb * 1.38.6 2024-02-10 [1] Bioconductor 3.18 (R 4.3.2)
GenomeInfoDbData 1.2.11 2024-02-27 [1] Bioconductor
GenomicAlignments 1.38.2 2024-01-20 [1] Bioconductor 3.18 (R 4.3.2)
GenomicFeatures * 1.54.3 2024-02-03 [1] Bioconductor 3.18 (R 4.3.2)
GenomicRanges * 1.54.1 2023-10-30 [1] Bioconductor
getPass 0.2-4 2023-12-10 [1] CRAN (R 4.3.1)
ggbeeswarm 0.7.2 2023-04-29 [1] CRAN (R 4.3.0)
ggplot2 * 3.5.0 2024-02-23 [1] CRAN (R 4.3.1)
ggrepel 0.9.5 2024-01-10 [1] CRAN (R 4.3.1)
ggridges 0.5.6 2024-01-23 [1] CRAN (R 4.3.1)
git2r 0.33.0 2023-11-26 [1] CRAN (R 4.3.1)
globals 0.16.2 2022-11-21 [1] CRAN (R 4.3.0)
glue * 1.7.0 2024-01-09 [1] CRAN (R 4.3.1)
GO.db * 3.18.0 2024-02-27 [1] Bioconductor
goftest 1.2-3 2021-10-07 [1] CRAN (R 4.3.0)
graph 1.80.0 2023-10-26 [1] Bioconductor
gridExtra 2.3 2017-09-09 [1] CRAN (R 4.3.0)
gtable 0.3.4 2023-08-21 [1] CRAN (R 4.3.0)
here * 1.0.1 2020-12-13 [1] CRAN (R 4.3.0)
highr 0.10 2022-12-22 [1] CRAN (R 4.3.0)
hms 1.1.3 2023-03-21 [1] CRAN (R 4.3.0)
Homo.sapiens * 1.3.1 2024-02-27 [1] Bioconductor
htmltools 0.5.7 2023-11-03 [1] CRAN (R 4.3.1)
htmlwidgets 1.6.4 2023-12-06 [1] CRAN (R 4.3.1)
httpuv 1.6.14 2024-01-26 [1] CRAN (R 4.3.1)
httr 1.4.7 2023-08-15 [1] CRAN (R 4.3.0)
ica 1.0-3 2022-07-08 [1] CRAN (R 4.3.0)
igraph 2.0.2 2024-02-17 [1] CRAN (R 4.3.1)
IRanges * 2.36.0 2023-10-26 [1] Bioconductor
irlba 2.3.5.1 2022-10-03 [1] CRAN (R 4.3.2)
janitor * 2.2.0 2023-02-02 [1] CRAN (R 4.3.0)
jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.3.0)
jsonlite 1.8.8 2023-12-04 [1] CRAN (R 4.3.1)
KEGGREST 1.42.0 2023-10-26 [1] Bioconductor
KernSmooth 2.23-22 2023-07-10 [1] CRAN (R 4.3.2)
knitr 1.45 2023-10-30 [1] CRAN (R 4.3.1)
labeling 0.4.3 2023-08-29 [1] CRAN (R 4.3.0)
later 1.3.2 2023-12-06 [1] CRAN (R 4.3.1)
lattice 0.22-5 2023-10-24 [1] CRAN (R 4.3.1)
lazyeval 0.2.2 2019-03-15 [1] CRAN (R 4.3.0)
leiden 0.4.3.1 2023-11-17 [1] CRAN (R 4.3.1)
lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.3.1)
limma 3.58.1 2023-11-02 [1] Bioconductor
listenv 0.9.1 2024-01-29 [1] CRAN (R 4.3.1)
lmtest 0.9-40 2022-03-21 [1] CRAN (R 4.3.0)
locfit 1.5-9.8 2023-06-11 [1] CRAN (R 4.3.0)
lubridate * 1.9.3 2023-09-27 [1] CRAN (R 4.3.1)
magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.0)
MASS 7.3-60.0.1 2024-01-13 [1] CRAN (R 4.3.1)
Matrix 1.6-5 2024-01-11 [1] CRAN (R 4.3.1)
MatrixGenerics * 1.14.0 2023-10-26 [1] Bioconductor
matrixStats * 1.2.0 2023-12-11 [1] CRAN (R 4.3.1)
memoise 2.0.1 2021-11-26 [1] CRAN (R 4.3.0)
metapod 1.10.1 2023-12-23 [1] Bioconductor 3.18 (R 4.3.2)
mime 0.12 2021-09-28 [1] CRAN (R 4.3.0)
miniUI 0.1.1.1 2018-05-18 [1] CRAN (R 4.3.0)
msigdbr * 7.5.1 2022-03-30 [1] CRAN (R 4.3.0)
munsell 0.5.0 2018-06-12 [1] CRAN (R 4.3.0)
nlme 3.1-164 2023-11-27 [1] CRAN (R 4.3.1)
org.Hs.eg.db * 3.18.0 2024-02-27 [1] Bioconductor
OrganismDbi * 1.44.0 2023-10-26 [1] Bioconductor
parallelly 1.37.0 2024-02-14 [1] CRAN (R 4.3.1)
patchwork * 1.2.0 2024-01-08 [1] CRAN (R 4.3.1)
pbapply 1.7-2 2023-06-27 [1] CRAN (R 4.3.0)
pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.0)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.0)
plotly 4.10.4 2024-01-13 [1] CRAN (R 4.3.1)
plyr 1.8.9 2023-10-02 [1] CRAN (R 4.3.1)
png 0.1-8 2022-11-29 [1] CRAN (R 4.3.0)
polyclip 1.10-6 2023-09-27 [1] CRAN (R 4.3.1)
prettyunits 1.2.0 2023-09-24 [1] CRAN (R 4.3.1)
processx 3.8.3 2023-12-10 [1] CRAN (R 4.3.1)
progress 1.2.3 2023-12-06 [1] CRAN (R 4.3.1)
progressr 0.14.0 2023-08-10 [1] CRAN (R 4.3.0)
promises 1.2.1 2023-08-10 [1] CRAN (R 4.3.0)
ProtGenerics 1.34.0 2023-10-26 [1] Bioconductor
ps 1.7.6 2024-01-18 [1] CRAN (R 4.3.1)
purrr * 1.0.2 2023-08-10 [1] CRAN (R 4.3.0)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.0)
RANN 2.6.1 2019-01-08 [1] CRAN (R 4.3.0)
rappdirs 0.3.3 2021-01-31 [1] CRAN (R 4.3.0)
RBGL 1.78.0 2023-10-26 [1] Bioconductor
RColorBrewer * 1.1-3 2022-04-03 [1] CRAN (R 4.3.0)
Rcpp 1.0.12 2024-01-09 [1] CRAN (R 4.3.1)
RcppAnnoy 0.0.22 2024-01-23 [1] CRAN (R 4.3.1)
RcppHNSW 0.6.0 2024-02-04 [1] CRAN (R 4.3.1)
RCurl 1.98-1.14 2024-01-09 [1] CRAN (R 4.3.1)
readr * 2.1.5 2024-01-10 [1] CRAN (R 4.3.1)
reshape2 1.4.4 2020-04-09 [1] CRAN (R 4.3.0)
restfulr 0.0.15 2022-06-16 [1] CRAN (R 4.3.0)
reticulate 1.35.0 2024-01-31 [1] CRAN (R 4.3.1)
rjson 0.2.21 2022-01-09 [1] CRAN (R 4.3.0)
rlang 1.1.3 2024-01-10 [1] CRAN (R 4.3.1)
rmarkdown 2.25 2023-09-18 [1] CRAN (R 4.3.1)
ROCR 1.0-11 2020-05-02 [1] CRAN (R 4.3.0)
rprojroot 2.0.4 2023-11-05 [1] CRAN (R 4.3.1)
Rsamtools 2.18.0 2023-10-26 [1] Bioconductor
RSpectra 0.16-1 2022-04-24 [1] CRAN (R 4.3.0)
RSQLite 2.3.5 2024-01-21 [1] CRAN (R 4.3.1)
rstudioapi 0.15.0 2023-07-07 [1] CRAN (R 4.3.0)
rsvd 1.0.5 2021-04-16 [1] CRAN (R 4.3.0)
rtracklayer 1.62.0 2023-10-26 [1] Bioconductor
Rtsne 0.17 2023-12-07 [1] CRAN (R 4.3.1)
S4Arrays 1.2.0 2023-10-26 [1] Bioconductor
S4Vectors * 0.40.2 2023-11-25 [1] Bioconductor 3.18 (R 4.3.2)
sass 0.4.8 2023-12-06 [1] CRAN (R 4.3.1)
ScaledMatrix 1.10.0 2023-11-06 [1] Bioconductor
scales * 1.3.0 2023-11-28 [1] CRAN (R 4.3.1)
scater * 1.30.1 2023-11-16 [1] Bioconductor
scattermore 1.2 2023-06-12 [1] CRAN (R 4.3.0)
scran * 1.30.2 2024-01-23 [1] Bioconductor 3.18 (R 4.3.2)
sctransform 0.4.1 2023-10-19 [1] CRAN (R 4.3.1)
scuttle * 1.12.0 2023-11-06 [1] Bioconductor
sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
Seurat * 5.0.1.9009 2024-02-28 [1] Github (satijalab/seurat@6a3ef5e)
SeuratObject * 5.0.1 2023-11-17 [1] CRAN (R 4.3.1)
shiny 1.8.0 2023-11-17 [1] CRAN (R 4.3.1)
SingleCellExperiment * 1.24.0 2023-11-06 [1] Bioconductor
snakecase 0.11.1 2023-08-27 [1] CRAN (R 4.3.0)
sp * 2.1-3 2024-01-30 [1] CRAN (R 4.3.1)
spam 2.10-0 2023-10-23 [1] CRAN (R 4.3.1)
SparseArray 1.2.4 2024-02-10 [1] Bioconductor 3.18 (R 4.3.2)
sparseMatrixStats 1.14.0 2023-10-26 [1] Bioconductor
spatstat.data 3.0-4 2024-01-15 [1] CRAN (R 4.3.1)
spatstat.explore 3.2-6 2024-02-01 [1] CRAN (R 4.3.1)
spatstat.geom 3.2-8 2024-01-26 [1] CRAN (R 4.3.1)
spatstat.random 3.2-2 2023-11-29 [1] CRAN (R 4.3.1)
spatstat.sparse 3.0-3 2023-10-24 [1] CRAN (R 4.3.1)
spatstat.utils 3.0-4 2023-10-24 [1] CRAN (R 4.3.1)
statmod 1.5.0 2023-01-06 [1] CRAN (R 4.3.0)
stringi 1.8.3 2023-12-11 [1] CRAN (R 4.3.1)
stringr * 1.5.1 2023-11-14 [1] CRAN (R 4.3.1)
SummarizedExperiment * 1.32.0 2023-11-06 [1] Bioconductor
survival 3.5-8 2024-02-14 [1] CRAN (R 4.3.1)
tensor 1.5 2012-05-05 [1] CRAN (R 4.3.0)
tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.3.0)
tidyr * 1.3.1 2024-01-24 [1] CRAN (R 4.3.1)
tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.3.0)
tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.3.0)
timechange 0.3.0 2024-01-18 [1] CRAN (R 4.3.1)
TxDb.Hsapiens.UCSC.hg19.knownGene * 3.2.2 2024-02-27 [1] Bioconductor
tzdb 0.4.0 2023-05-12 [1] CRAN (R 4.3.0)
utf8 1.2.4 2023-10-22 [1] CRAN (R 4.3.1)
uwot 0.1.16 2023-06-29 [1] CRAN (R 4.3.0)
vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.3.1)
vipor 0.4.7 2023-12-18 [1] CRAN (R 4.3.1)
viridis 0.6.5 2024-01-29 [1] CRAN (R 4.3.1)
viridisLite 0.4.2 2023-05-02 [1] CRAN (R 4.3.0)
whisker 0.4.1 2022-12-05 [1] CRAN (R 4.3.0)
withr 3.0.0 2024-01-16 [1] CRAN (R 4.3.1)
workflowr * 1.7.1 2023-08-23 [1] CRAN (R 4.3.0)
xfun 0.42 2024-02-08 [1] CRAN (R 4.3.1)
XML 3.99-0.16.1 2024-01-22 [1] CRAN (R 4.3.1)
xml2 1.3.6 2023-12-04 [1] CRAN (R 4.3.1)
xtable 1.8-4 2019-04-21 [1] CRAN (R 4.3.0)
XVector 0.42.0 2023-10-26 [1] Bioconductor
yaml 2.3.8 2023-12-11 [1] CRAN (R 4.3.1)
zlibbioc 1.48.0 2023-10-26 [1] Bioconductor
zoo 1.8-12 2023-04-13 [1] CRAN (R 4.3.0)
[1] /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/library
──────────────────────────────────────────────────────────────────────────────
sessionInfo()R version 4.3.2 (2023-10-31)
Platform: aarch64-apple-darwin20 (64-bit)
Running under: macOS Sonoma 14.5
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: Australia/Melbourne
tzcode source: internal
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] EnsDb.Hsapiens.v86_2.99.0
[2] ensembldb_2.26.0
[3] AnnotationFilter_1.26.0
[4] msigdbr_7.5.1
[5] Homo.sapiens_1.3.1
[6] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
[7] org.Hs.eg.db_3.18.0
[8] GO.db_3.18.0
[9] OrganismDbi_1.44.0
[10] GenomicFeatures_1.54.3
[11] AnnotationDbi_1.64.1
[12] scales_1.3.0
[13] patchwork_1.2.0
[14] cowplot_1.1.3
[15] janitor_2.2.0
[16] Seurat_5.0.1.9009
[17] SeuratObject_5.0.1
[18] sp_2.1-3
[19] scater_1.30.1
[20] scran_1.30.2
[21] scuttle_1.12.0
[22] SingleCellExperiment_1.24.0
[23] SummarizedExperiment_1.32.0
[24] Biobase_2.62.0
[25] GenomicRanges_1.54.1
[26] GenomeInfoDb_1.38.6
[27] IRanges_2.36.0
[28] S4Vectors_0.40.2
[29] BiocGenerics_0.48.1
[30] MatrixGenerics_1.14.0
[31] matrixStats_1.2.0
[32] RColorBrewer_1.1-3
[33] glue_1.7.0
[34] here_1.0.1
[35] lubridate_1.9.3
[36] forcats_1.0.0
[37] stringr_1.5.1
[38] dplyr_1.1.4
[39] purrr_1.0.2
[40] readr_2.1.5
[41] tidyr_1.3.1
[42] tibble_3.2.1
[43] ggplot2_3.5.0
[44] tidyverse_2.0.0
[45] BiocStyle_2.30.0
[46] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] ProtGenerics_1.34.0 fs_1.6.3
[3] spatstat.sparse_3.0-3 bitops_1.0-7
[5] httr_1.4.7 tools_4.3.2
[7] sctransform_0.4.1 utf8_1.2.4
[9] R6_2.5.1 lazyeval_0.2.2
[11] uwot_0.1.16 withr_3.0.0
[13] prettyunits_1.2.0 gridExtra_2.3
[15] progressr_0.14.0 cli_3.6.2
[17] spatstat.explore_3.2-6 fastDummies_1.7.3
[19] labeling_0.4.3 sass_0.4.8
[21] spatstat.data_3.0-4 ggridges_0.5.6
[23] pbapply_1.7-2 Rsamtools_2.18.0
[25] sessioninfo_1.2.2 parallelly_1.37.0
[27] limma_3.58.1 rstudioapi_0.15.0
[29] RSQLite_2.3.5 BiocIO_1.12.0
[31] generics_0.1.3 ica_1.0-3
[33] spatstat.random_3.2-2 Matrix_1.6-5
[35] ggbeeswarm_0.7.2 fansi_1.0.6
[37] abind_1.4-5 lifecycle_1.0.4
[39] whisker_0.4.1 yaml_2.3.8
[41] edgeR_4.0.16 snakecase_0.11.1
[43] BiocFileCache_2.10.1 SparseArray_1.2.4
[45] Rtsne_0.17 grid_4.3.2
[47] blob_1.2.4 promises_1.2.1
[49] dqrng_0.3.2 crayon_1.5.2
[51] miniUI_0.1.1.1 lattice_0.22-5
[53] beachmat_2.18.1 KEGGREST_1.42.0
[55] pillar_1.9.0 knitr_1.45
[57] metapod_1.10.1 rjson_0.2.21
[59] future.apply_1.11.1 codetools_0.2-19
[61] leiden_0.4.3.1 getPass_0.2-4
[63] data.table_1.15.0 vctrs_0.6.5
[65] png_0.1-8 spam_2.10-0
[67] gtable_0.3.4 cachem_1.0.8
[69] xfun_0.42 S4Arrays_1.2.0
[71] mime_0.12 survival_3.5-8
[73] statmod_1.5.0 bluster_1.12.0
[75] ellipsis_0.3.2 fitdistrplus_1.1-11
[77] ROCR_1.0-11 nlme_3.1-164
[79] bit64_4.0.5 filelock_1.0.3
[81] progress_1.2.3 RcppAnnoy_0.0.22
[83] rprojroot_2.0.4 bslib_0.6.1
[85] irlba_2.3.5.1 vipor_0.4.7
[87] KernSmooth_2.23-22 colorspace_2.1-0
[89] DBI_1.2.2 tidyselect_1.2.0
[91] processx_3.8.3 curl_5.2.0
[93] bit_4.0.5 compiler_4.3.2
[95] git2r_0.33.0 graph_1.80.0
[97] BiocNeighbors_1.20.2 xml2_1.3.6
[99] DelayedArray_0.28.0 plotly_4.10.4
[101] rtracklayer_1.62.0 lmtest_0.9-40
[103] RBGL_1.78.0 callr_3.7.5
[105] rappdirs_0.3.3 digest_0.6.34
[107] goftest_1.2-3 spatstat.utils_3.0-4
[109] rmarkdown_2.25 XVector_0.42.0
[111] htmltools_0.5.7 pkgconfig_2.0.3
[113] sparseMatrixStats_1.14.0 highr_0.10
[115] dbplyr_2.4.0 fastmap_1.1.1
[117] rlang_1.1.3 htmlwidgets_1.6.4
[119] shiny_1.8.0 DelayedMatrixStats_1.24.0
[121] farver_2.1.1 jquerylib_0.1.4
[123] zoo_1.8-12 jsonlite_1.8.8
[125] BiocParallel_1.36.0 BiocSingular_1.18.0
[127] RCurl_1.98-1.14 magrittr_2.0.3
[129] GenomeInfoDbData_1.2.11 dotCall64_1.1-1
[131] munsell_0.5.0 Rcpp_1.0.12
[133] babelgene_22.9 viridis_0.6.5
[135] reticulate_1.35.0 stringi_1.8.3
[137] zlibbioc_1.48.0 MASS_7.3-60.0.1
[139] plyr_1.8.9 parallel_4.3.2
[141] listenv_0.9.1 ggrepel_0.9.5
[143] deldir_2.0-2 Biostrings_2.70.2
[145] splines_4.3.2 tensor_1.5
[147] hms_1.1.3 locfit_1.5-9.8
[149] ps_1.7.6 igraph_2.0.2
[151] spatstat.geom_3.2-8 RcppHNSW_0.6.0
[153] biomaRt_2.58.2 reshape2_1.4.4
[155] ScaledMatrix_1.10.0 XML_3.99-0.16.1
[157] evaluate_0.23 BiocManager_1.30.22
[159] tzdb_0.4.0 httpuv_1.6.14
[161] RANN_2.6.1 polyclip_1.10-6
[163] future_1.33.1 scattermore_1.2
[165] rsvd_1.0.5 xtable_1.8-4
[167] restfulr_0.0.15 RSpectra_0.16-1
[169] later_1.3.2 viridisLite_0.4.2
[171] GenomicAlignments_1.38.2 memoise_2.0.1
[173] beeswarm_0.4.0 cluster_2.1.6
[175] timechange_0.3.0 globals_0.16.2