Master_metadata
Combined plots for all batches
Gunjan Dixit
February 04, 2025
Last updated: 2025-02-04
Checks: 5 2
Knit directory: paed-airway-allTissues/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R
Markdown file created these results, you’ll want to first commit it to
the Git repo. If you’re still working on the analysis, you can ignore
this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230811) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| ~/projects/paed-airway-atlas/airway-atlas-allTissues/paed-airway-allTissues/output/RDS/AllBatches_Annotation_SEUs_v2 | output/RDS/AllBatches_Annotation_SEUs_v2 |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 54e4ec2. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: data/.DS_Store
Ignored: data/RDS/
Ignored: output/.DS_Store
Ignored: output/CSV/.DS_Store
Ignored: output/G000231_Neeland_batch1/
Ignored: output/G000231_Neeland_batch2_1/
Ignored: output/G000231_Neeland_batch2_2/
Ignored: output/G000231_Neeland_batch3/
Ignored: output/G000231_Neeland_batch4/
Ignored: output/G000231_Neeland_batch5/
Ignored: output/G000231_Neeland_batch9_1/
Ignored: output/RDS/
Ignored: output/plots/
Untracked files:
Untracked: analysis/03_Batch_Integration.Rmd
Untracked: analysis/Age_proportions.Rmd
Untracked: analysis/Age_proportions_AllBatches.Rmd
Untracked: analysis/All_Batches_QCExploratory_v2.Rmd
Untracked: analysis/All_metadata.Rmd
Untracked: analysis/Annotation_BAL.Rmd
Untracked: analysis/Annotation_Bronchial_brushings.Rmd
Untracked: analysis/Annotation_Nasal_brushings.Rmd
Untracked: analysis/BatchCorrection_Adenoids.Rmd
Untracked: analysis/BatchCorrection_Nasal_brushings.Rmd
Untracked: analysis/BatchCorrection_Tonsils.Rmd
Untracked: analysis/Batch_Integration_&_Downstream_analysis.Rmd
Untracked: analysis/Batch_correction_&_Downstream.Rmd
Untracked: analysis/Cell_cycle_regression.Rmd
Untracked: analysis/Clustering_Tonsils_v2.Rmd
Untracked: analysis/DGE_analysis_George.Rmd
Untracked: analysis/Master_metadata.Rmd
Untracked: analysis/Pediatric_Vs_Adult_Atlases.Rmd
Untracked: analysis/Preprocessing_Batch1_Nasal_brushings.Rmd
Untracked: analysis/Preprocessing_Batch2_Tonsils.Rmd
Untracked: analysis/Preprocessing_Batch3_Adenoids.Rmd
Untracked: analysis/Preprocessing_Batch4_Bronchial_brushings.Rmd
Untracked: analysis/Preprocessing_Batch5_Nasal_brushings.Rmd
Untracked: analysis/Preprocessing_Batch6_BAL.Rmd
Untracked: analysis/Preprocessing_Batch7_Bronchial_brushings.Rmd
Untracked: analysis/Preprocessing_Batch8_Adenoids.Rmd
Untracked: analysis/Preprocessing_Batch9_Tonsils.Rmd
Untracked: analysis/TonsilsVsAdenoids.Rmd
Untracked: analysis/cell_cycle_regression.R
Untracked: analysis/testing_age_all.Rmd
Untracked: data/Cell_labels_Gunjan_v2/
Untracked: data/Cell_labels_Mel/
Untracked: data/Cell_labels_Mel_v2/
Untracked: data/Cell_labels_Mel_v3/
Untracked: data/Cell_labels_modified_Gunjan/
Untracked: data/Gene_sets/
Untracked: data/Hs.c2.cp.reactome.v7.1.entrez.rds
Untracked: data/Raw_feature_bc_matrix/
Untracked: data/cell_labels_Mel_v4_Dec2024/
Untracked: data/celltypes_Mel_GD_v3.xlsx
Untracked: data/celltypes_Mel_GD_v4_no_dups.xlsx
Untracked: data/celltypes_Mel_modified.xlsx
Untracked: data/celltypes_Mel_v2.csv
Untracked: data/celltypes_Mel_v2.xlsx
Untracked: data/celltypes_Mel_v2_MN.xlsx
Untracked: data/celltypes_for_mel_MN.xlsx
Untracked: data/col_palette.xlsx
Untracked: data/earlyAIR_sample_sheets_combined.xlsx
Untracked: data/~$col_palette.xlsx
Untracked: output/CSV/All_tissues.propeller.xlsx
Untracked: output/CSV/Bronchial_brushings/
Untracked: output/CSV/Bronchial_brushings_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/
Untracked: output/CSV/G000231_Neeland_Adenoids.propeller.xlsx
Untracked: output/CSV/G000231_Neeland_Bronchial_brushings.propeller.xlsx
Untracked: output/CSV/G000231_Neeland_Nasal_brushings.propeller.xlsx
Untracked: output/CSV/G000231_Neeland_Tonsils.propeller.xlsx
Untracked: output/CSV/Nasal_brushings/
Untracked: output/CSV_v2/G000231_Neeland_Adenoids.propeller.xlsx
Untracked: output/CSV_v2/G000231_Neeland_Nasal_brushings.propeller.xlsx
Untracked: output/CSV_v2/G000231_Neeland_Tonsils.propeller.xlsx
Untracked: output/DGE/
Untracked: test_col.csv
Untracked: test_col.txt
Untracked: test_col.xlsx
Unstaged changes:
Deleted: 02_QC_exploratoryPlots.Rmd
Deleted: 02_QC_exploratoryPlots.html
Modified: analysis/00_AllBatches_overview.Rmd
Modified: analysis/01_QC_emptyDrops.Rmd
Modified: analysis/02_QC_exploratoryPlots.Rmd
Modified: analysis/Adenoids.Rmd
Modified: analysis/Adenoids_v2.Rmd
Modified: analysis/Age_modeling.Rmd
Modified: analysis/Age_modelling_Adenoids.Rmd
Modified: analysis/Age_modelling_Nasal_Brushings.Rmd
Modified: analysis/Age_modelling_Tonsils.Rmd
Modified: analysis/AllBatches_QCExploratory.Rmd
Modified: analysis/BAL.Rmd
Modified: analysis/BAL_v2.Rmd
Modified: analysis/Bronchial_brushings.Rmd
Modified: analysis/Bronchial_brushings_v2.Rmd
Modified: analysis/Nasal_brushings.Rmd
Modified: analysis/Nasal_brushings_v2.Rmd
Modified: analysis/Subclustering_Adenoids.Rmd
Modified: analysis/Subclustering_BAL.Rmd
Modified: analysis/Subclustering_Bronchial_brushings.Rmd
Modified: analysis/Subclustering_Nasal_brushings.Rmd
Modified: analysis/Subclustering_Tonsils.Rmd
Modified: analysis/Tonsils.Rmd
Modified: analysis/Tonsils_v2.Rmd
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c0.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c1.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c10.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c11.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c12.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c13.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c14.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c15.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c16.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c17.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c2.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c3.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c4.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c5.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c6.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c7.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c8.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c9.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c0.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c1.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c10.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c11.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c12.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c13.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c14.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c15.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c16.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c17.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c2.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c3.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c4.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c5.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c6.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c7.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c8.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c9.csv
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with
wflow_publish() to start tracking its development.
Load libraries
suppressPackageStartupMessages({
library(BiocStyle)
library(tidyverse)
library(here)
library(dplyr)
library(Seurat)
library(clustree)
library(paletteer)
library(viridis)
library(ggforce)
library(ggridges)
library(kableExtra)
library(RColorBrewer)
library(data.table)
library(dplyr)
library(cowplot)
library(ggplot2)
library(paletteer)
library(patchwork)
library(harmony)
library(BiocParallel)
library(circlize)
})data_path <- here("~/projects/paed-airway-atlas/airway-atlas-allTissues/paed-airway-allTissues/output/RDS/AllBatches_Annotation_SEUs_v2")
tissue_list <- list.files(data_path, pattern = "\\.rds$", full.names = TRUE)
tissue_list[1] "/Users/dixitgunjan/projects/paed-airway-atlas/airway-atlas-allTissues/paed-airway-allTissues/output/RDS/AllBatches_Annotation_SEUs_v2/G000231_Neeland_Adenoids.annotated_clusters.SEU.rds"
[2] "/Users/dixitgunjan/projects/paed-airway-atlas/airway-atlas-allTissues/paed-airway-allTissues/output/RDS/AllBatches_Annotation_SEUs_v2/G000231_Neeland_BAL.annotated_clusters.SEU.rds"
[3] "/Users/dixitgunjan/projects/paed-airway-atlas/airway-atlas-allTissues/paed-airway-allTissues/output/RDS/AllBatches_Annotation_SEUs_v2/G000231_Neeland_Bronchial_brushings.annotated_clusters.SEU.rds"
[4] "/Users/dixitgunjan/projects/paed-airway-atlas/airway-atlas-allTissues/paed-airway-allTissues/output/RDS/AllBatches_Annotation_SEUs_v2/G000231_Neeland_Nasal_brushings.annotated_clusters.SEU.rds"
[5] "/Users/dixitgunjan/projects/paed-airway-atlas/airway-atlas-allTissues/paed-airway-allTissues/output/RDS/AllBatches_Annotation_SEUs_v2/G000231_Neeland_Tonsils.annotated_clusters.SEU.rds" metadata_list <- list()
for (tissue in tissue_list) {
seu <- readRDS(tissue)
metadata <- seu@meta.data
metadata$source <- basename(tissue)
metadata_list[[tissue]] <- metadata
}
combined_metadata <- bind_rows(metadata_list)sort(table(combined_metadata$cell_labels_v2), decreasing = T)
Naïve B cells Memory B cells
87050 51442
Macrophages Non-ciliated epithelial cells
37383 34992
CD4 TFH Ciliated epithelial cells
25963 23752
CD4 TN DZtoLZ GCB transition
17887 17762
TFH-LZ-GC Naïve B cell-IFN
17170 14607
Plasma B cells DZ G2Mphase
14373 13881
Early MBC DZ GCB
13055 12961
CD8 TRM DZ early Sphase
12317 10212
CD8 TF Monocyte and neutrophil-like
9481 9318
DZ late Sphase CD8 TN
8618 8398
CD4 Treg-eff Early GC-committed NBC
8293 8086
T-IFN Cycling GCB
7501 7288
Naïve B cells activated CD4 TCM
6632 6562
Monocytes/macrophages Neutrophils
5481 5095
Intermediate B cells CD4 TEM
4749 4479
CD8 TEM IFN-activated cells
3909 3687
GC-commited metabolic activation Monocytes
3613 3548
Pre-BCRi II NK cells
3452 3352
Unconventional T cells Plasmacytoid DCs
3331 3316
CD4 effector Follicular dendritic cells
2944 2811
Dendritic cells Macro-CCL
2494 2434
CD4 Treg B activated
2296 2045
Double negative T Early PC precursor
2026 2020
Mast cells Macro-proliferating
1877 1813
Proliferating epithelial cells Gamma delta T
1799 1657
Pre-MBC/BC NK/gamma-delta T
1651 1546
Secretory epithelial cells CD4 T proliferating
1142 1086
Proliferating B cells Proliferating T/NK
1086 1075
csMBC FCRL4/5+ Pre-T cells
960 937
NK-T cells Erythroid cells
832 802
Epithelial cells B cells
779 726
DZ GCB Noproli-memory like MAIT cells
694 603
Ionocytes Cycling T
558 536
CD8 T GCB-IFN
387 324
Basal epithelial cells Melanocyte
206 205
Mesothelial cells
199 unique(combined_metadata$cell_labels_v2) [1] "Naïve B cells" "Memory B cells"
[3] "Plasma B cells" "Naïve B cell-IFN"
[5] "Monocytes/macrophages" "Follicular dendritic cells"
[7] "Pre-BCRi II" "Plasmacytoid DCs"
[9] "Epithelial cells" "Mast cells"
[11] "Neutrophils" "DZtoLZ GCB transition"
[13] "DZ early Sphase" "Early MBC"
[15] "Early GC-committed NBC" "DZ G2Mphase"
[17] "DZ GCB Noproli-memory like" "DZ GCB"
[19] "DZ late Sphase" "GC-commited metabolic activation"
[21] "csMBC FCRL4/5+" "Cycling GCB"
[23] "Early PC precursor" "Pre-T cells"
[25] "CD4 T proliferating" "CD4 TN"
[27] "CD4 TFH" "T-IFN"
[29] "CD4 TCM" "NK-T cells"
[31] "CD4 Treg-eff" "TFH-LZ-GC"
[33] "CD8 TF" "CD8 TN"
[35] "NK/gamma-delta T" "Double negative T"
[37] "B cells" "Basal epithelial cells"
[39] "Macrophages" "Macro-proliferating"
[41] "Secretory epithelial cells" "Dendritic cells"
[43] "Ciliated epithelial cells" "Macro-CCL"
[45] "Monocytes" "Pre-MBC/BC"
[47] "Proliferating B cells" "B activated"
[49] "CD4 TEM" "CD4 Treg"
[51] "CD8 TRM" "CD8 TEM"
[53] "NK cells" "CD8 T"
[55] "Proliferating T/NK" "MAIT cells"
[57] "Mesothelial cells" "Monocyte and neutrophil-like"
[59] "Non-ciliated epithelial cells" "Ionocytes"
[61] "Intermediate B cells" "Unconventional T cells"
[63] "Melanocyte" "Proliferating epithelial cells"
[65] "IFN-activated cells" "Erythroid cells"
[67] "Naïve B cells activated" "GCB-IFN"
[69] "CD4 effector" "Gamma delta T"
[71] "Cycling T" unique(combined_metadata$Broad_cell_label_3) [1] "B cells" "Dendritic cells"
[3] "Macrophages" "Doublet query/Other"
[5] "Epithelial lineage" "Granulocytes"
[7] "CD4 T cells" "Gamma delta T cells"
[9] "Double negative T cells" "CD8 T cells"
[11] "Pre B/T cells" "Cycling T cells"
[13] "Innate lymphoid cells" "Natural Killer cells"
[15] "Other" "Monocytes"
[17] "Neuroendocrine" "SMG duct"
[19] "Fibroblast lineage" "Endothelial lineage" Exploratory figures
To see cell counts across all Tissues
cell_type_counts <- sort(table(combined_metadata$cell_labels_v2), decreasing = TRUE) %>%
as.data.frame() %>%
rename(CellType = Var1, Count = Freq)
a <- ggplot(cell_type_counts, aes(x = reorder(CellType, Count), y = Count)) +
geom_bar(stat = "identity", fill = "purple3") +
geom_text(aes(label = Count), hjust = -0.1, size = 3) + # Position the text just outside the bar
coord_flip() +
labs(title = "Cell Type counts in earlyAIR Atlas", x = "Cell Types", y = "Cell Count") +
theme_minimal()
a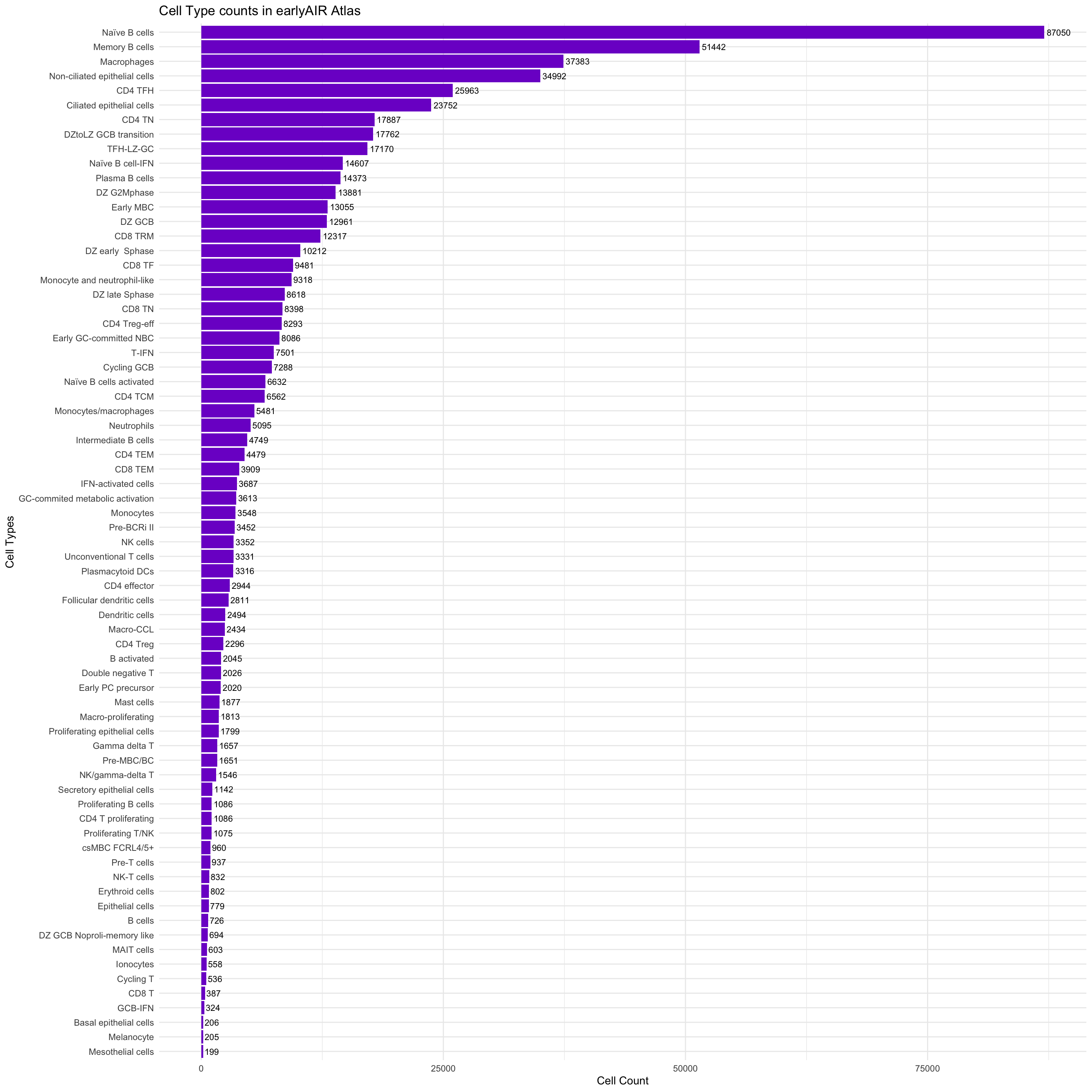
cell_type_tissue_counts <- combined_metadata %>%
group_by(cell_labels_v2, tissue) %>%
summarise(Count = n(), .groups = 'drop') %>%
rename(CellType = cell_labels_v2)
total_counts <- cell_type_tissue_counts %>%
group_by(CellType) %>%
summarise(TotalCount = sum(Count)) %>%
arrange(desc(TotalCount))
cell_type_tissue_counts$CellType <- factor(cell_type_tissue_counts$CellType, levels = rev(total_counts$CellType))
ggplot(cell_type_tissue_counts, aes(x = CellType, y = Count, fill = tissue)) +
geom_bar(stat = "identity") +
geom_text(data = total_counts, aes(x = CellType, y = TotalCount, label = TotalCount),
hjust = -0.1, size = 3, inherit.aes = FALSE) +
coord_flip() +
scale_fill_brewer(palette = "Set3") +
labs(title = "Cell Type Distribution by Tissue in earlyAIR Atlas", x = "Cell Types", y = "Cell Count") +
theme_minimal() +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust = 1))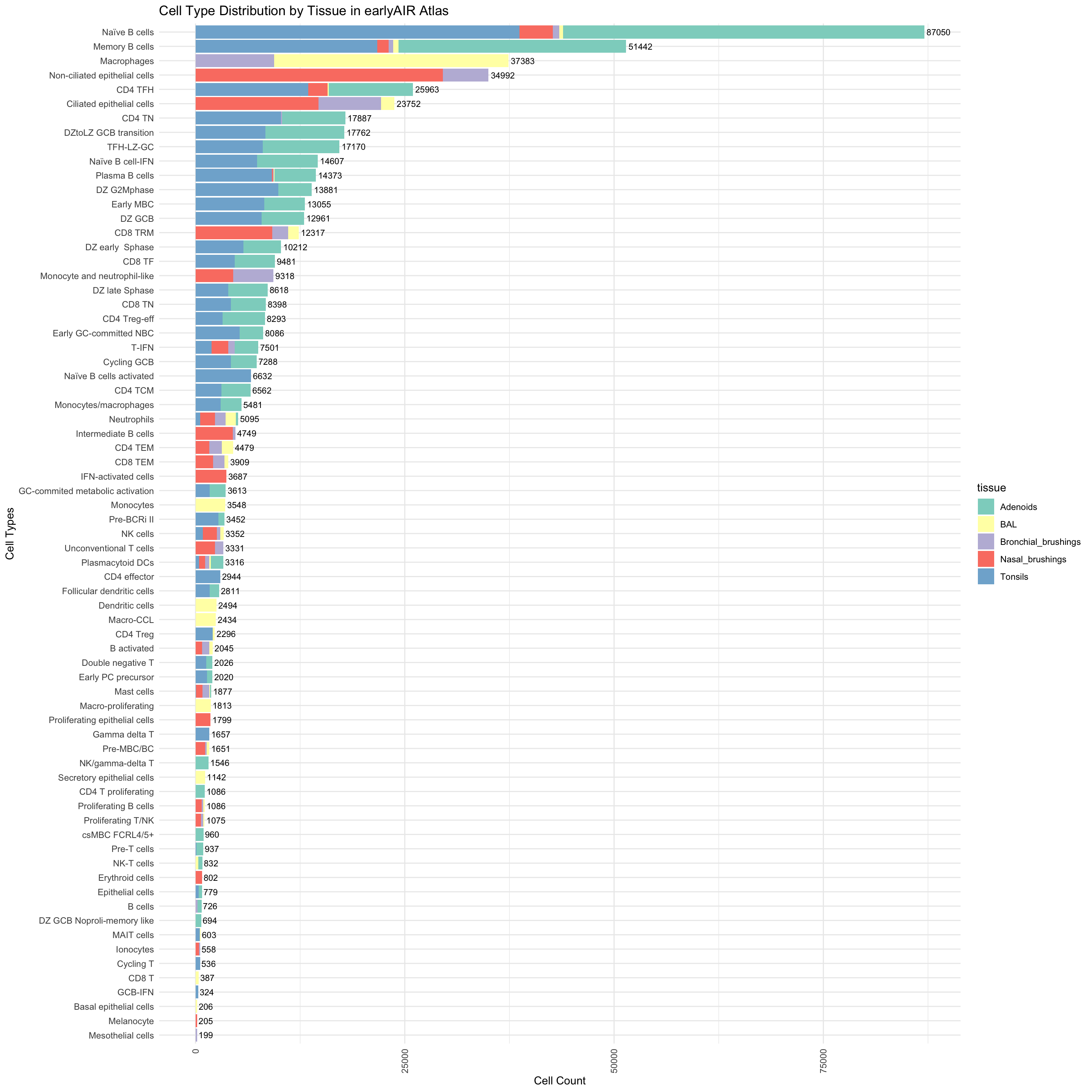
ggplot(cell_type_tissue_counts, aes(x = CellType, y = Count, fill = tissue)) +
geom_bar(stat = "identity") +
coord_flip() +
geom_text(aes(label = Count), position = position_stack(vjust = 0.5), size = 2.5) +
scale_fill_brewer(palette = "Set3") + # Customize palette
labs(title = "Cell Type Distribution by Tissue", x = "Cell Types", y = "Cell Count", fill = "Tissue") +
theme_minimal() +
theme(axis.text.y = element_text(size = 8))ggplot(combined_metadata, aes(x = cell_labels_v2, fill = tissue)) +
geom_bar(position = "dodge") +
facet_wrap(~tissue, scales = "free_y") +
coord_flip() +
labs(title = "Cell Type Counts by Tissue", x = "Cell Types", y = "Count") +
theme_minimal()
ggplot(combined_metadata, aes(x = tissue, fill = cell_labels_v2)) +
geom_bar(position = "stack") +
geom_text(stat = "count", aes(label = ..count..), position = position_stack(vjust = 0.5), size = 2.5) +
labs(title = "Cell Type Counts per Tissue", x = "Tissue", y = "Count") +
theme_minimal()#set.seed(012025)
#n <- 71
n <- length(unique(combined_metadata$cell_labels_v2))
qual_col_pals <- brewer.pal.info[brewer.pal.info$category == 'qual',]
col_vector <- unlist(mapply(brewer.pal, qual_col_pals$maxcolors, rownames(qual_col_pals)))
sampled_colors <- sample(col_vector, n, replace = TRUE)
cell_types <- unique(combined_metadata$cell_labels_v2)
#color_palette <- setNames(sampled_colors[1:length(cell_types)], cell_types)
color_palette <- readRDS(here("output/RDS/color_palette_unique.rds"))
proportion_df <- combined_metadata %>%
group_by(tissue, cell_labels_v2) %>%
summarise(Count = n()) %>%
mutate(Proportion = Count / sum(Count))`summarise()` has grouped output by 'tissue'. You can override using the
`.groups` argument.sampled_colors_1 <- c("#5c248b", "#1f57a6", "#ffec34", "#00960f", "#BC80BD", "#f06ab9", "#85d519", "#758dc4", "#89c5df", "#5da3cd", "#ffffba", "#009260", "#ffa037", "#A65628", "#E31A1C", "#377EB8" ,"#FDC086", "#FC8D62" ,"#FDDAEC", "#E78AC3")
proportion_df <- combined_metadata %>%
group_by(tissue, Broad_cell_label_3) %>%
summarise(Count = n()) %>%
mutate(Proportion = Count / sum(Count))`summarise()` has grouped output by 'tissue'. You can override using the
`.groups` argument.tissue_order <- c("Tonsils", "Adenoids", "Nasal_brushings", "Bronchial_brushings", "BAL")
proportion_df$tissue <- factor(proportion_df$tissue, levels = tissue_order)
p_stacked <- ggplot(proportion_df, aes(x = tissue, y = Proportion, fill = Broad_cell_label_3)) +
geom_bar(stat = "identity") +
scale_fill_manual(values = sampled_colors_1) +
ylab("Proportion of Cell Labels") +
theme_cowplot(font_size = 10) +
labs(title = "Proportion of Cell Labels per Tissue") +
theme(axis.text.x = element_text(angle = 45, hjust = 1),
plot.title = element_text(hjust = 0.5)) # Center the title
print(p_stacked)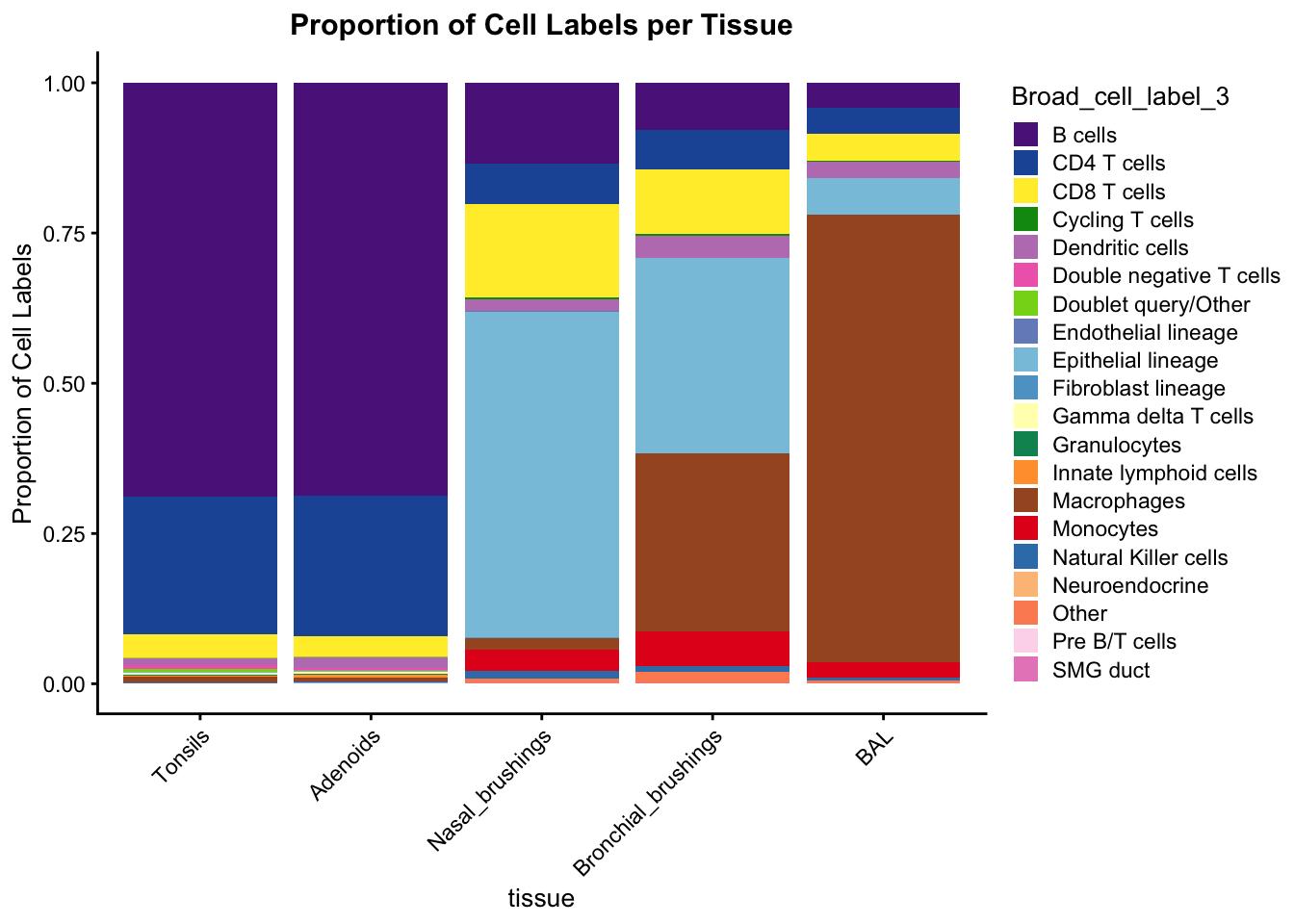
combined_metadata <- combined_metadata %>%
mutate(Broad_category = case_when(
# B Cells
cell_labels_v2 %in% c("Naïve B cells", "Memory B cells", "Naïve B cell-IFN",
"Plasma B cells", "DZtoLZ GCB transition", "DZ early Sphase",
"Early MBC", "Early GC-committed NBC", "DZ G2Mphase",
"DZ GCB Noproli-memory like", "DZ GCB", "DZ late Sphase",
"GC-commited metabolic activation", "csMBC FCRL4/5+",
"Cycling GCB", "Early PC precursor", "Pre-MBC/BC",
"Proliferating B cells", "B cells", "Intermediate B cells",
"Naïve B cells activated", "Pre-BCRi II", "IFN-activated cells",
"GCB-IFN", "B activated") ~ "B Cells",
# T Cells
cell_labels_v2 %in% c("CD4 TN", "CD4 TFH", "CD4 TCM", "CD4 Treg",
"CD4 Treg-eff", "TFH-LZ-GC", "CD4 TEM", "CD4 effector",
"CD4 T proliferating", "T-IFN", "CD8 TN", "CD8 TF",
"CD8 T", "CD8 TRM", "CD8 TEM", "Double negative T",
"Unconventional T cells", "Gamma delta T", "Cycling T", "Pre-T cells") ~ "T Cells",
# NK Cells
cell_labels_v2 %in% c("NK cells", "NK-T cells", "NK/gamma-delta T",
"Proliferating T/NK") ~ "NK Cells",
# Monocytes and Macrophages
cell_labels_v2 %in% c("Monocytes", "Monocytes/macrophages", "Macrophages",
"Macro-proliferating", "Macro-CCL", "Monocyte and neutrophil-like") ~ "Monocytes and Macrophages",
# Dendritic Cells
cell_labels_v2 %in% c("Dendritic cells", "Plasmacytoid DCs", "Follicular dendritic cells") ~ "Dendritic Cells",
# Neutrophils
cell_labels_v2 == "Neutrophils" ~ "Neutrophils",
# Innate Lymphoid Cells
cell_labels_v2 %in% c("MAIT cells", "Innate lymphocytes") ~ "Innate Lymphoid Cells",
# Epithelial Cells
cell_labels_v2 %in% c("Epithelial cells", "Basal epithelial cells", "Ciliated epithelial cells",
"Non-ciliated epithelial cells", "Secretory epithelial cells",
"Proliferating epithelial cells") ~ "Epithelial Cells",
# Other
cell_labels_v2 %in% c( "Mast cells", "Erythroid cells", "Ionocytes", "Mesothelial cells",
"Melanocyte",
"Naïve / PC/ doublet") ~ "Other",
TRUE ~ "Unclassified" # Default category for unmatched labels
))
head(combined_metadata) donor_id sample_id age_years sex nCount_RNA
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 eAIR001 s042 3.62 M 2097.863
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 eAIR001 s042 3.62 M 4072.417
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 eAIR001 s042 3.62 M 2848.063
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 eAIR001 s042 3.62 M 434.372
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 eAIR001 s042 3.62 M 21590.274
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 eAIR001 s042 3.62 M 1144.299
nFeature_RNA Barcode
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 1486 AAACAAGCAACTTCGTACTTTAGG-1
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 2506 AAACAAGCATCGTTCGACTTTAGG-1
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 1891 AAACCAATCCTTTAGGACTTTAGG-1
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 443 AAACCGGTCCGTGACTACTTTAGG-1
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 4311 AAACGTTCAGCCCTTAACTTTAGG-1
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 877 AAACGTTCATGGCTAAACTTTAGG-1
GEM_barcode sample_barcode tissue
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 AAACAAGCAACTTCGTA CTTTAGG-1 Adenoids
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 AAACAAGCATCGTTCGA CTTTAGG-1 Adenoids
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 AAACCAATCCTTTAGGA CTTTAGG-1 Adenoids
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 AAACCGGTCCGTGACTA CTTTAGG-1 Adenoids
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 AAACGTTCAGCCCTTAA CTTTAGG-1 Adenoids
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 AAACGTTCATGGCTAAA CTTTAGG-1 Adenoids
batch_name cells_per_GEM.Var1
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 G000231_batch3 <NA>
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 G000231_batch3 <NA>
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 G000231_batch3 <NA>
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 G000231_batch3 <NA>
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 G000231_batch3 <NA>
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 G000231_batch3 <NA>
cells_per_GEM.Freq scDblFinder.class_dbr
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 3 singlet
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 3 singlet
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 2 singlet
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 1 singlet
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 3 singlet
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 1 singlet
scDblFinder.score_dbr predicted.celltype.l1
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 8.364258e-04 B naive
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 3.647992e-04 FCRL4/5+ B memory
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 6.277001e-03 B naive
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 6.357333e-07 B activated
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 3.589588e-02 PC
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 2.285886e-04 B naive
predicted.celltype.l2
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 NBC
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 ncsMBC
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 NBC early activation
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 GC-commited NBC
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 IgG+ PC precursor
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 NBC early activation
predicted.celltype.l1.score
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 0.9986553
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 0.5014055
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 0.9326043
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 0.5793129
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 0.8757269
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 0.9211898
predicted.celltype.l2.score percent.mt
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 0.5650053 0.89363848
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 0.3133930 0.45582175
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 0.4845545 0.45551262
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 0.5580569 0.37925250
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 0.6296915 0.06705518
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 0.8442881 0.60955238
mapping.score Broad_cell_label_1
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 0.8641857 Immune
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 0.7719604 Immune
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 0.7590004 Immune
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 0.8996206 Immune
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 0.8858244 Immune
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 0.8131308 Immune
Broad_cell_label_2 Broad_cell_label_3
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 B cells B cells
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 B cells B cells
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 B cells B cells
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 B cells B cells
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 B cells B cells
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 B cells B cells
unintegrated_clusters harmony_clusters
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 0 0
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 2 2
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 0 0
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 0 0
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 9 9
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 0 0
RNA_snn_res.0.1 RNA_snn_res.0.2
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 0 0
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 0 0
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 0 0
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 0 0
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 5 5
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 0 0
RNA_snn_res.0.3 RNA_snn_res.0.4
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 0 0
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 2 1
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 0 0
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 0 0
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 9 9
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 0 0
RNA_snn_res.0.5 RNA_snn_res.0.6
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 0 0
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 1 1
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 0 0
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 8 7
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 12 12
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 0 0
RNA_snn_res.0.7 RNA_snn_res.0.8
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 0 0
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 1 1
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 0 1
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 7 6
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 13 13
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 0 0
RNA_snn_res.0.9 RNA_snn_res.1 cluster
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 0 0 0
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 1 1 1
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 0 0 0
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 6 6 0
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 11 12 9
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 0 0 0
cell_labels cell_labels_v2
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 Naïve B cells Naïve B cells
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 Memory B cells Memory B cells
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 Naïve B cells Naïve B cells
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 Naïve B cells Naïve B cells
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 Plasma B cells Plasma B cells
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 Naïve B cells Naïve B cells
source
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 G000231_Neeland_Adenoids.annotated_clusters.SEU.rds
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 G000231_Neeland_Adenoids.annotated_clusters.SEU.rds
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 G000231_Neeland_Adenoids.annotated_clusters.SEU.rds
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 G000231_Neeland_Adenoids.annotated_clusters.SEU.rds
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 G000231_Neeland_Adenoids.annotated_clusters.SEU.rds
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 G000231_Neeland_Adenoids.annotated_clusters.SEU.rds
predicted.ann_level_1
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 <NA>
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 <NA>
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 <NA>
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 <NA>
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 <NA>
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 <NA>
predicted.ann_level_1.score
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 NA
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 NA
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 NA
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 NA
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 NA
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 NA
predicted.ann_level_2
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 <NA>
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 <NA>
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 <NA>
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 <NA>
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 <NA>
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 <NA>
predicted.ann_level_2.score
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 NA
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 NA
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 NA
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 NA
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 NA
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 NA
predicted.ann_level_3
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 <NA>
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 <NA>
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 <NA>
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 <NA>
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 <NA>
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 <NA>
predicted.ann_level_3.score
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 NA
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 NA
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 NA
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 NA
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 NA
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 NA
predicted.ann_level_4
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 <NA>
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 <NA>
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 <NA>
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 <NA>
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 <NA>
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 <NA>
predicted.ann_level_4.score
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 NA
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 NA
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 NA
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 NA
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 NA
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 NA
predicted.ann_level_5
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 <NA>
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 <NA>
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 <NA>
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 <NA>
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 <NA>
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 <NA>
predicted.ann_level_5.score
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 NA
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 NA
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 NA
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 NA
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 NA
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 NA
predicted.ann_finest_level
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 <NA>
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 <NA>
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 <NA>
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 <NA>
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 <NA>
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 <NA>
predicted.ann_finest_level.score donor sum
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 NA <NA> NA
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 NA <NA> NA
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 NA <NA> NA
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 NA <NA> NA
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 NA <NA> NA
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 NA <NA> NA
detected scDblFinder.class_dbr_s
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 NA <NA>
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 NA <NA>
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 NA <NA>
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 NA <NA>
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 NA <NA>
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 NA <NA>
scDblFinder.score_dbr_s Broad_category
Batch1_AAACAAGCAACTTCGTACTTTAGG-1 NA B Cells
Batch1_AAACAAGCATCGTTCGACTTTAGG-1 NA B Cells
Batch1_AAACCAATCCTTTAGGACTTTAGG-1 NA B Cells
Batch1_AAACCGGTCCGTGACTACTTTAGG-1 NA B Cells
Batch1_AAACGTTCAGCCCTTAACTTTAGG-1 NA B Cells
Batch1_AAACGTTCATGGCTAAACTTTAGG-1 NA B Cellscombined_metadata$Broad_category <- factor(combined_metadata$Broad_category,
levels = c("B Cells", "T Cells", "NK Cells", "Monocytes and Macrophages",
"Dendritic Cells", "Neutrophils", "Innate Lymphoid Cells",
"Epithelial Cells", "Other", "Unclassified"))
combined_metadata$tissue <- factor(combined_metadata$tissue, levels = tissue_order)
ggplot(combined_metadata, aes(x = tissue, fill = Broad_category)) +
geom_bar(position = "fill") +
scale_fill_manual(values = sampled_colors_1) +
labs(y = "Proportion", x = "Tissue") +
theme(axis.text.x = element_text(angle = 90, hjust = 1)) +
theme_minimal() +
theme(legend.title = element_blank()) 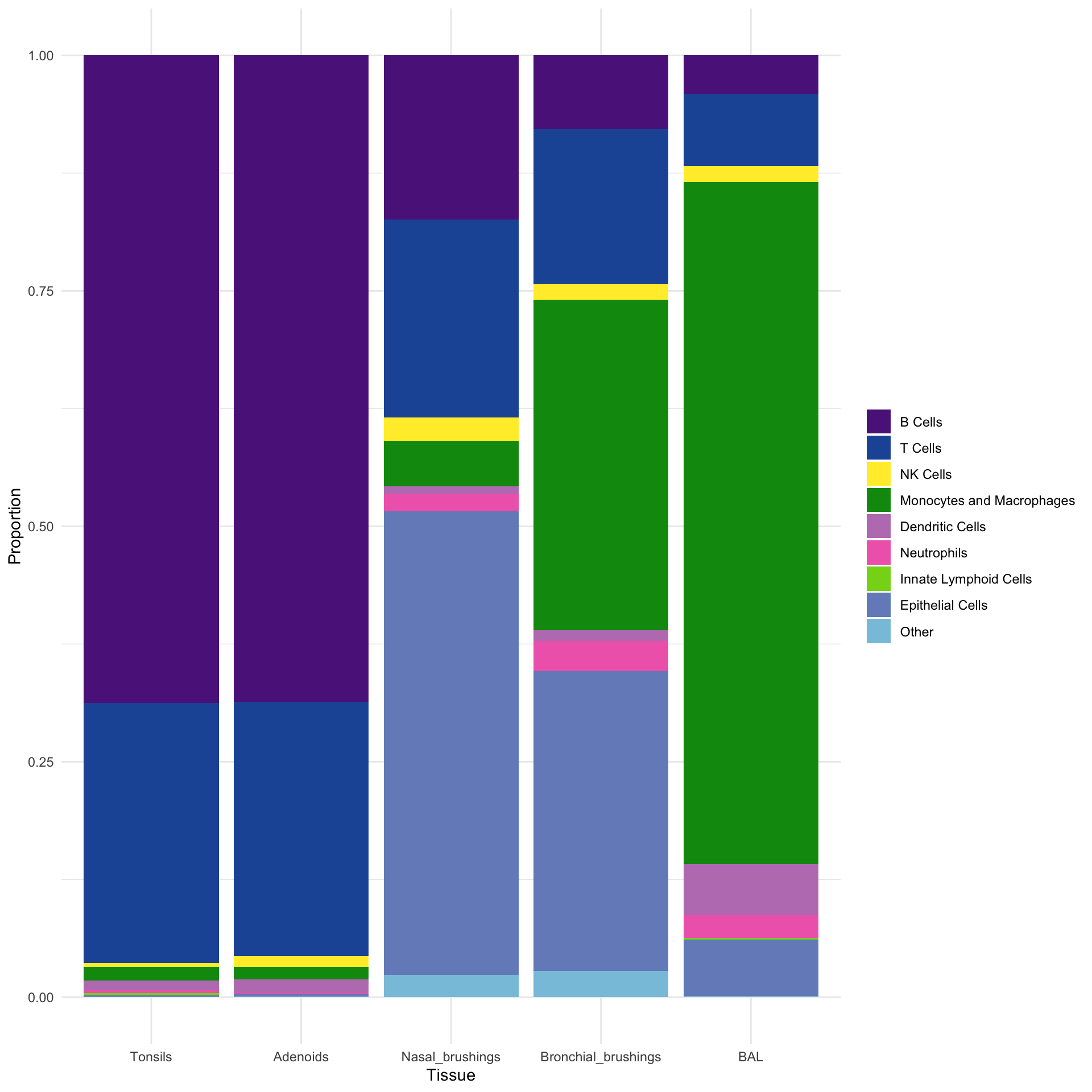
combined_metadata$cell_labels_v2 <- factor(combined_metadata$cell_labels_v2,
levels = unique(combined_metadata$cell_labels_v2))
ggplot(combined_metadata, aes(x = tissue, fill = cell_labels_v2)) +
geom_bar(position = "fill") +
scale_fill_manual(values = color_palette) +
labs(y = "Proportion", x = "Tissue") +
theme(axis.text.x = element_text(angle = 90, hjust = 1)) +
theme_minimal() +
theme(legend.title = element_blank()) 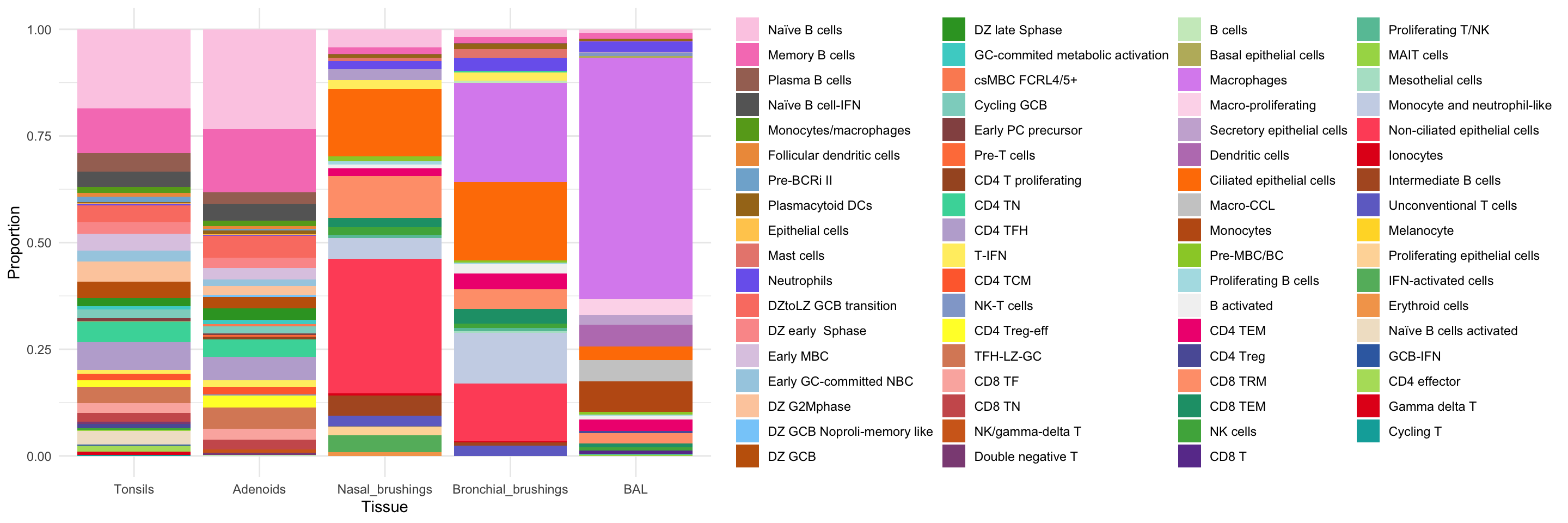
set.seed(2024)
proportions <- combined_metadata %>%
group_by(Broad_category, cell_labels_v2) %>%
summarise(count = n()) %>%
arrange(Broad_category, desc(count))`summarise()` has grouped output by 'Broad_category'. You can override using
the `.groups` argument.combined_metadata <- combined_metadata %>%
mutate(cell_labels_v2 = factor(cell_labels_v2,
levels = proportions$cell_labels_v2[order(proportions$Broad_category, -proportions$count)]))broad_colors <- c("#5c248b", "#ffec34", "#00960f", "#FBB4AE", "#BC80BD", "#f06ab9", "#85d519", "#758dc4" )
cell_contingency <- table(combined_metadata$Broad_category, combined_metadata$cell_labels_v2)
chordDiagram(cell_contingency,
transparency = 0.5,
annotationTrack = "grid",
preAllocateTracks = 1,
grid.col = c(broad_colors, color_palette))
circos.track(track.index = 1, panel.fun = function(x, y) {
circos.text(CELL_META$xcenter, CELL_META$ylim[1], CELL_META$sector.index,
facing = "clockwise", niceFacing = TRUE, adj = c(0, 0.5))
}, bg.border = NA)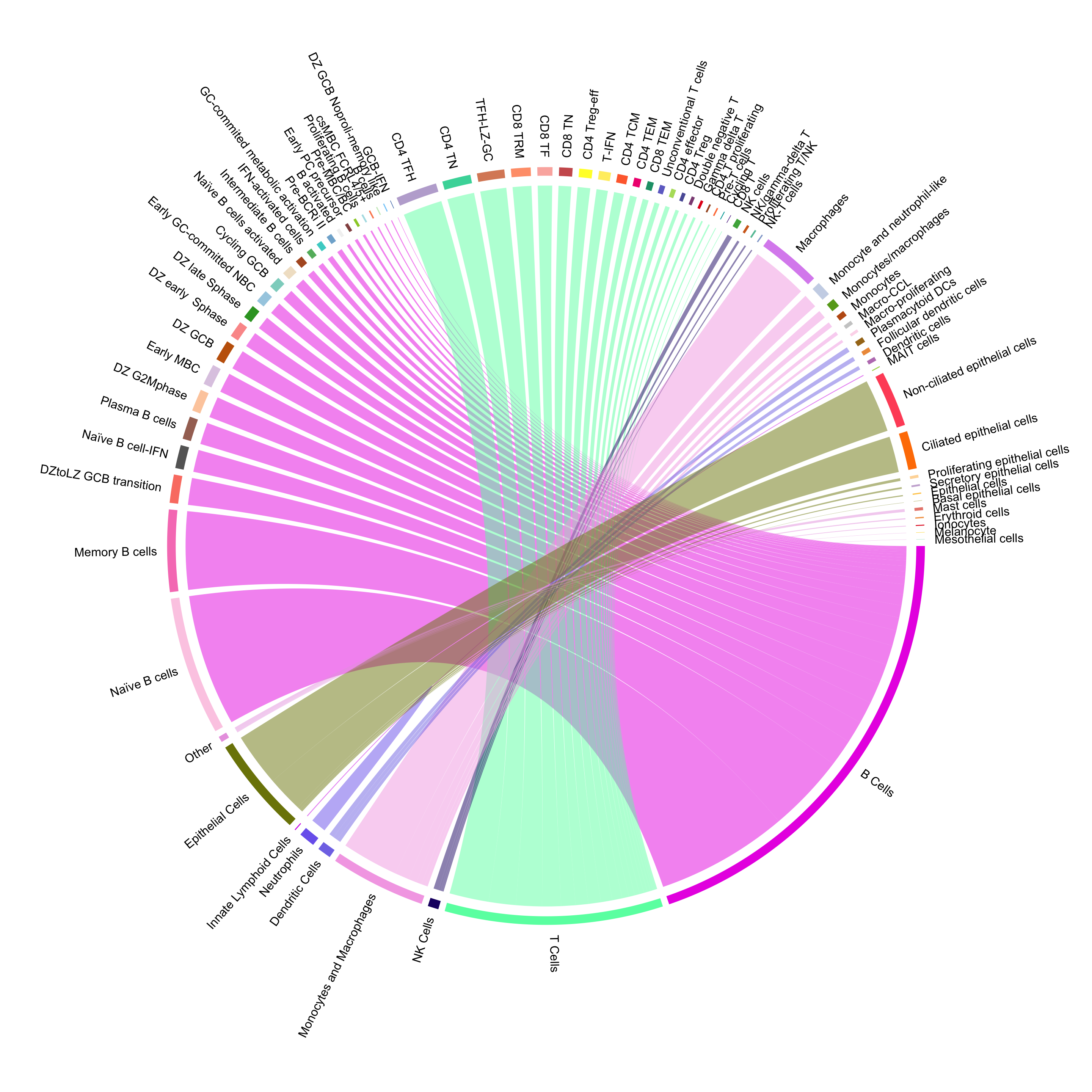
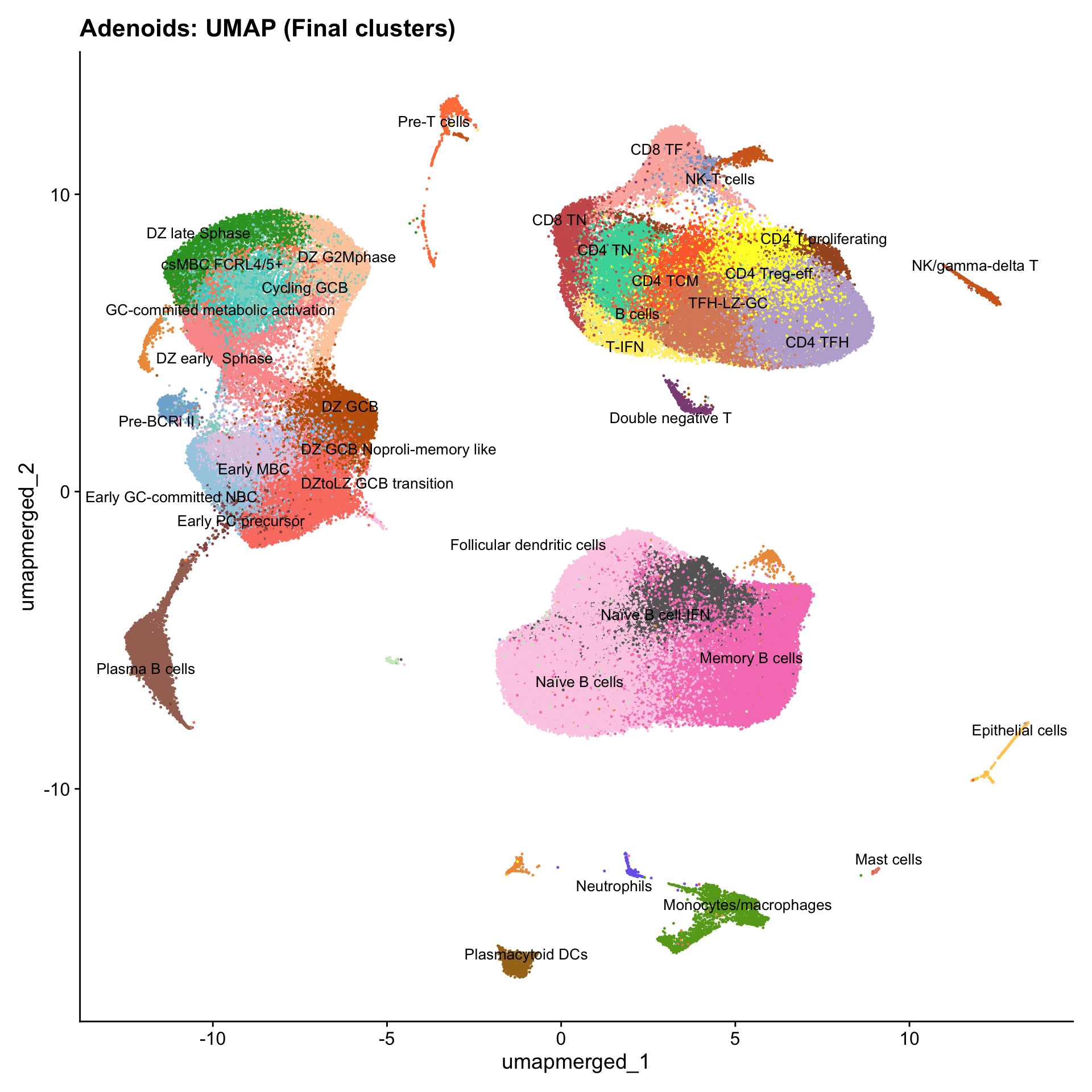
Updated UMAPs
for (tissue in tissue_list) {
seu <- readRDS(tissue)
tissue <- seu$tissue
p4 <- DimPlot(seu, reduction = "umap.merged", raster = FALSE, repel = TRUE, label = TRUE, label.size = 3.5, pt.size = 0.2) +
ggtitle(paste0(basename(tissue), ": UMAP (Final clusters)")) +
scale_color_manual(values = color_palette) +
NoLegend()
print(p4)
}
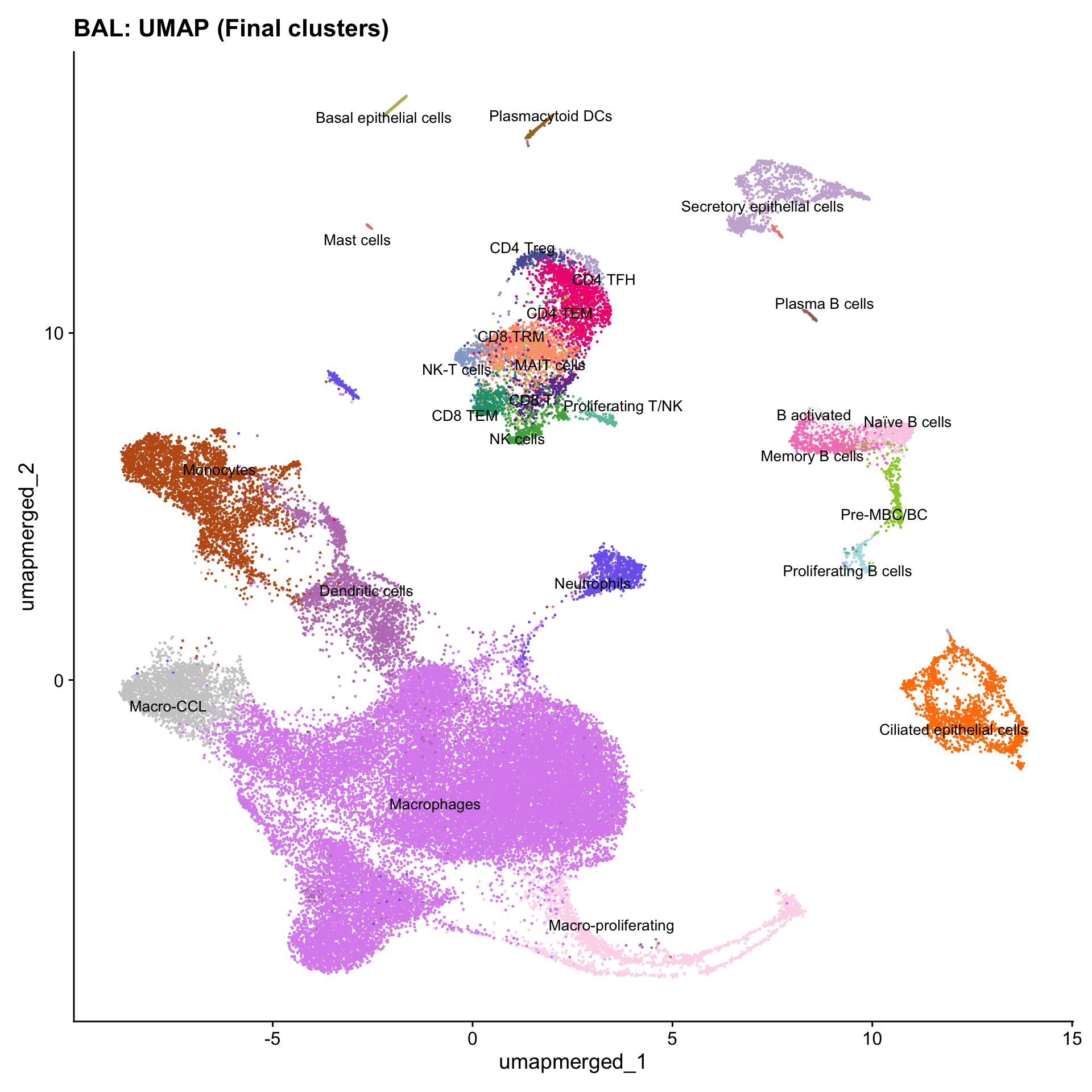
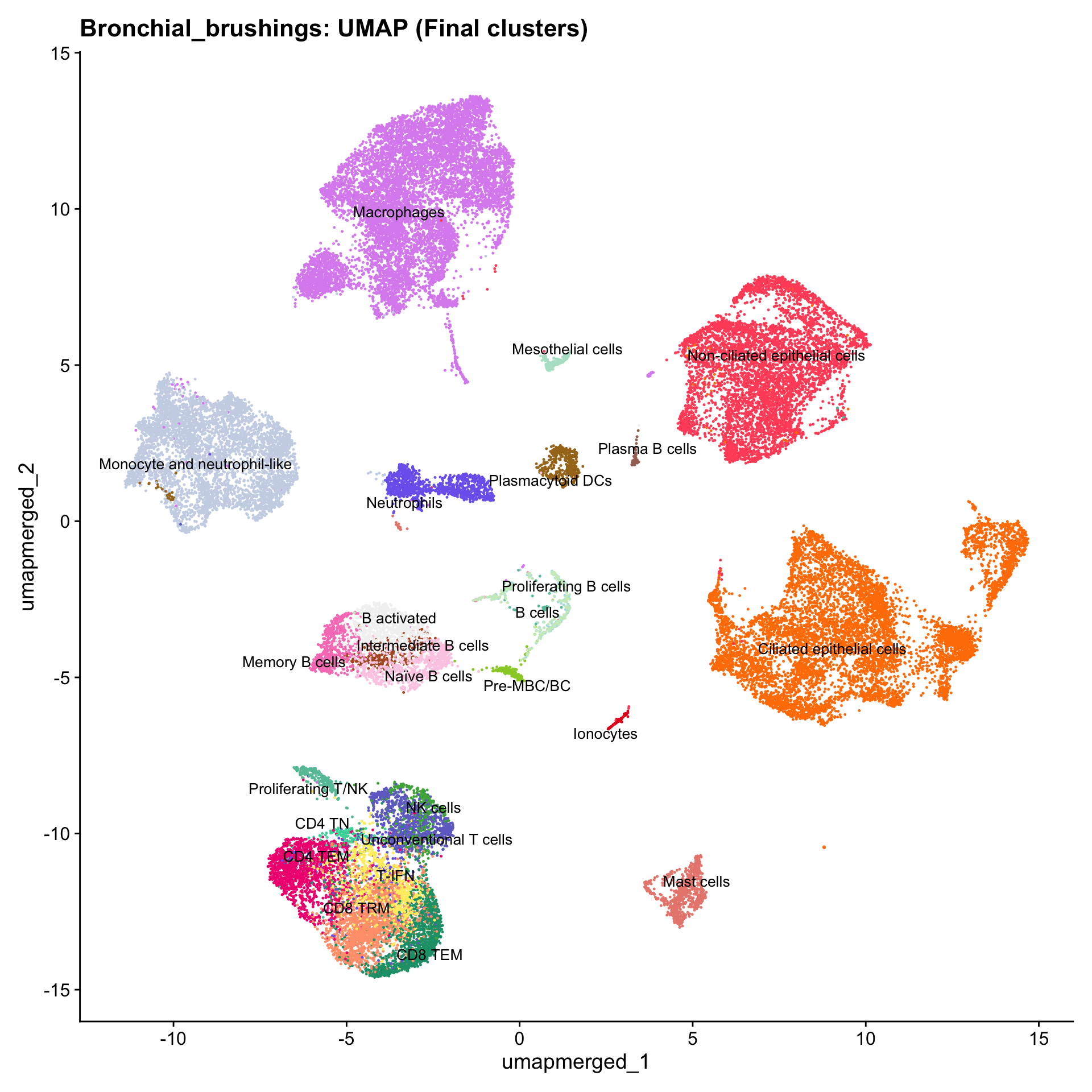
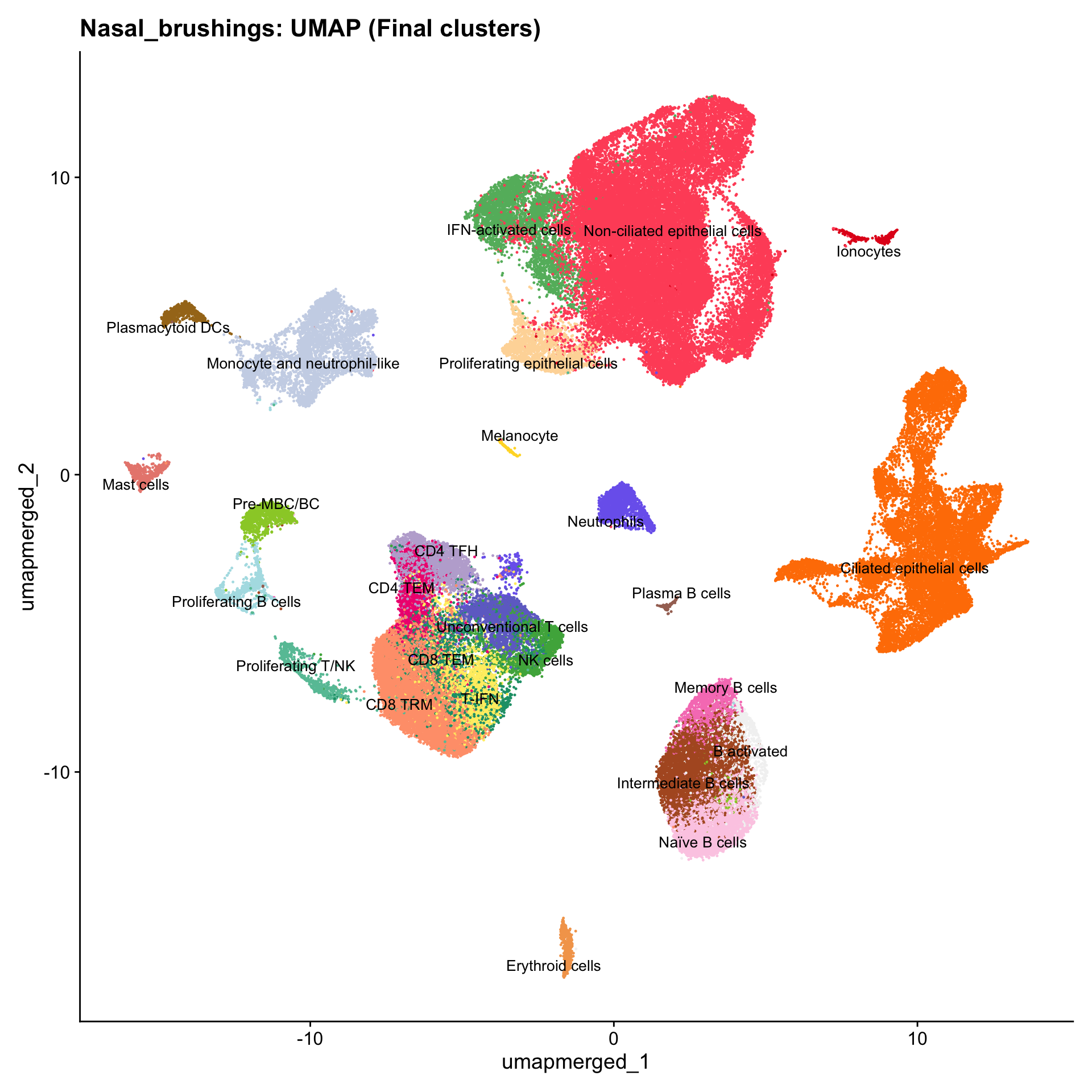
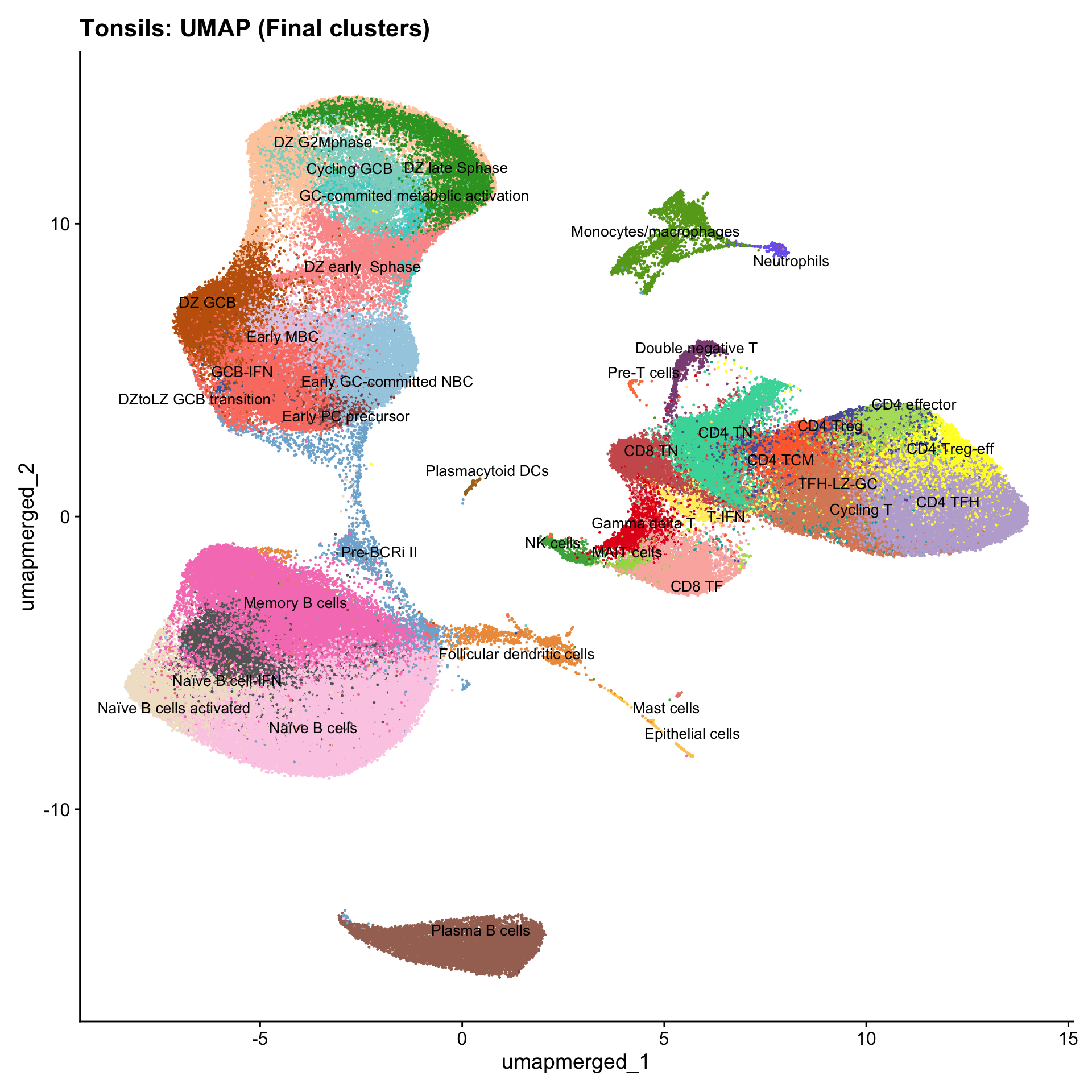
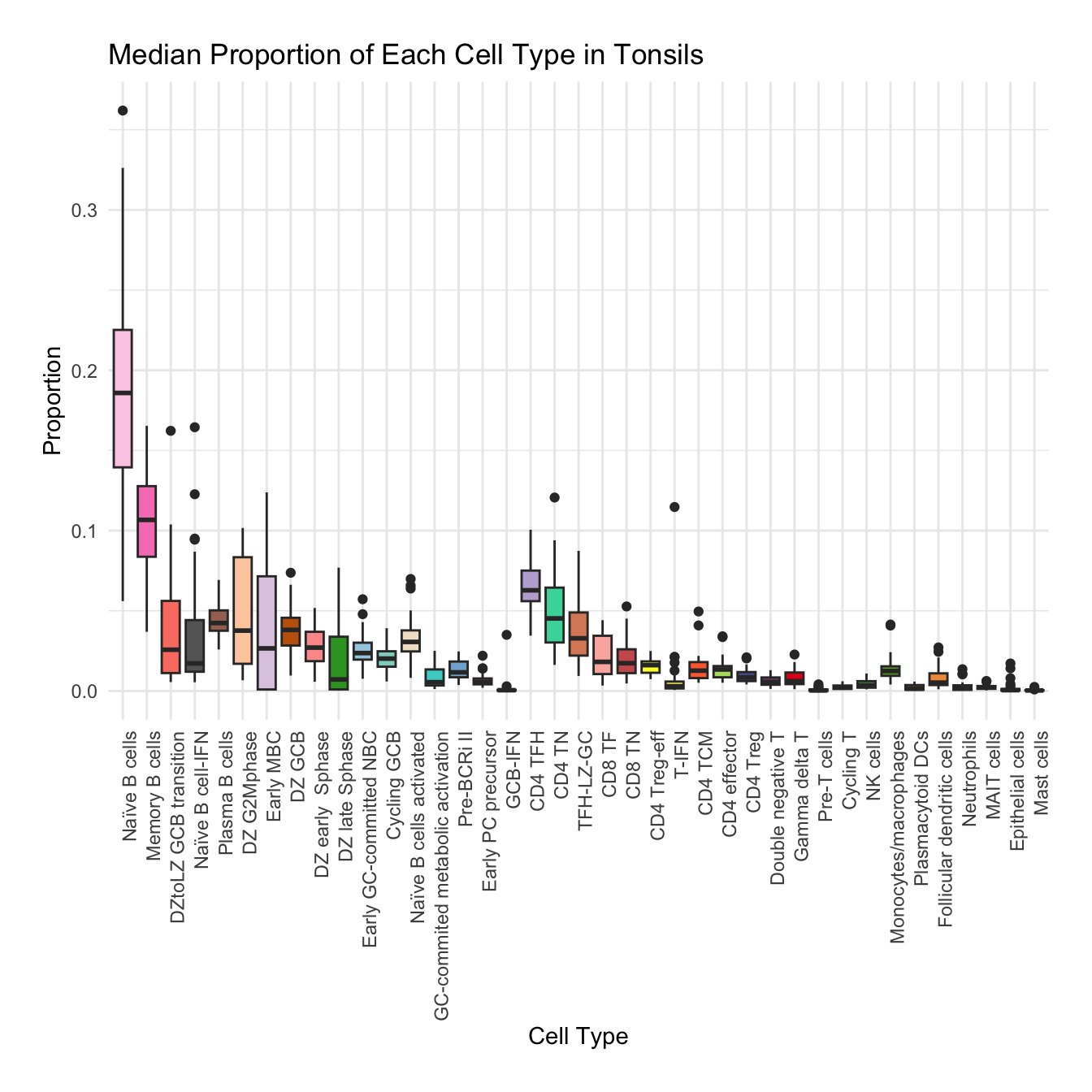
Boxplot of cell populations
cell_type_proportions <- combined_metadata %>%
group_by(tissue, sample_id, cell_labels_v2) %>%
summarise(cell_count = n(), .groups = 'drop') %>%
group_by(tissue, sample_id) %>%
mutate(total_cells = sum(cell_count)) %>%
mutate(proportion = cell_count / total_cells) %>%
ungroup()
tissues <- unique(cell_type_proportions$tissue)
for (tissue in tissues) {
tissue_data <- cell_type_proportions %>% filter(tissue == !!tissue)
plot <- ggplot(tissue_data, aes(x = cell_labels_v2, y = proportion, fill = cell_labels_v2)) +
geom_boxplot() +
scale_fill_manual(values = color_palette) +
labs(x = "Cell Type", y = "Proportion", title = paste("Median Proportion of Each Cell Type in", tissue)) +
theme_minimal() +
theme(
axis.text.x = element_text(angle = 90, hjust = 1),
legend.position = "none",
plot.margin = margin(20, 20, 20, 20)
)
#ggsave(filename = paste0("boxplot_proportions_", tissue, ".pdf"), plot = plot, width = 12, height = 8, units = "in")
print(plot)
}
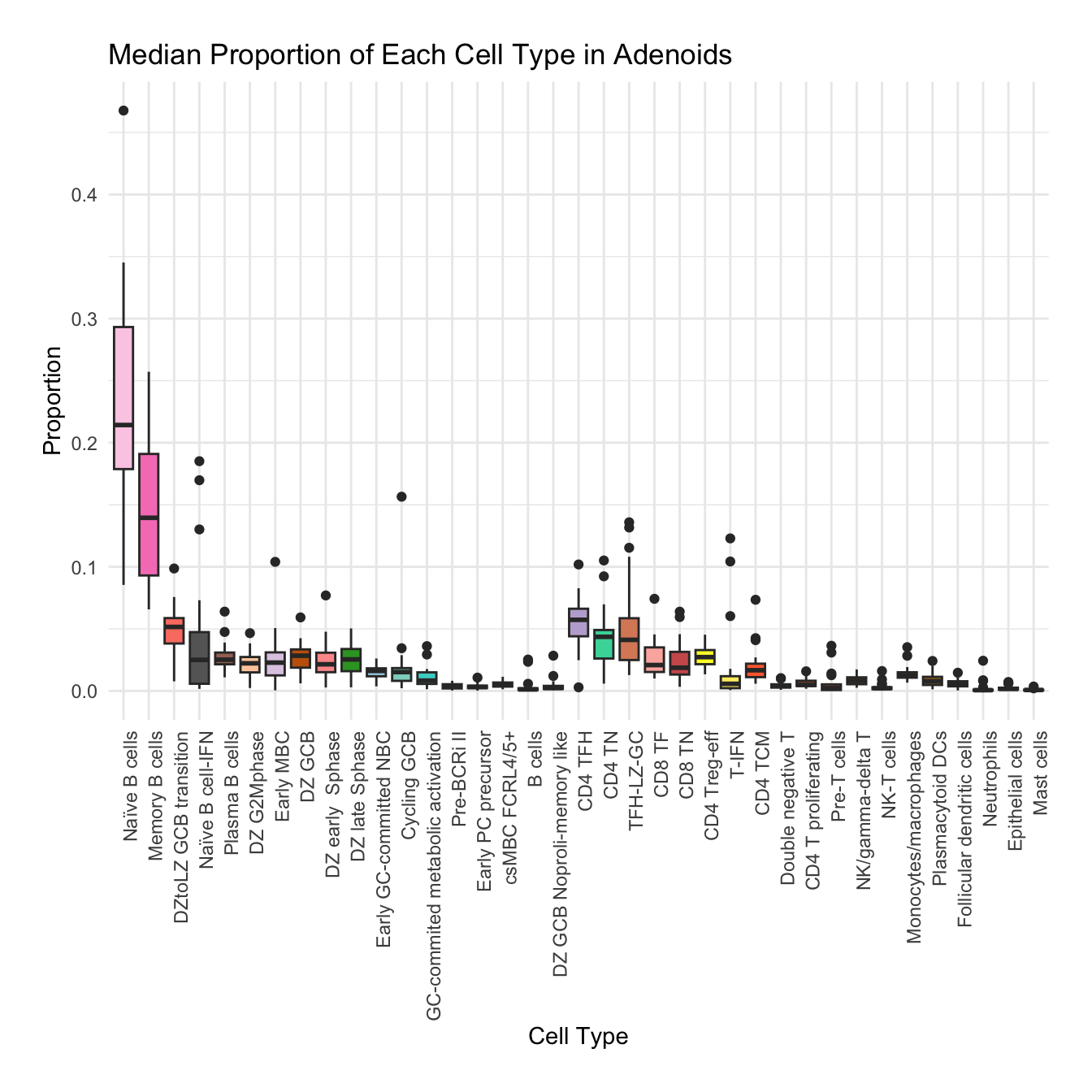
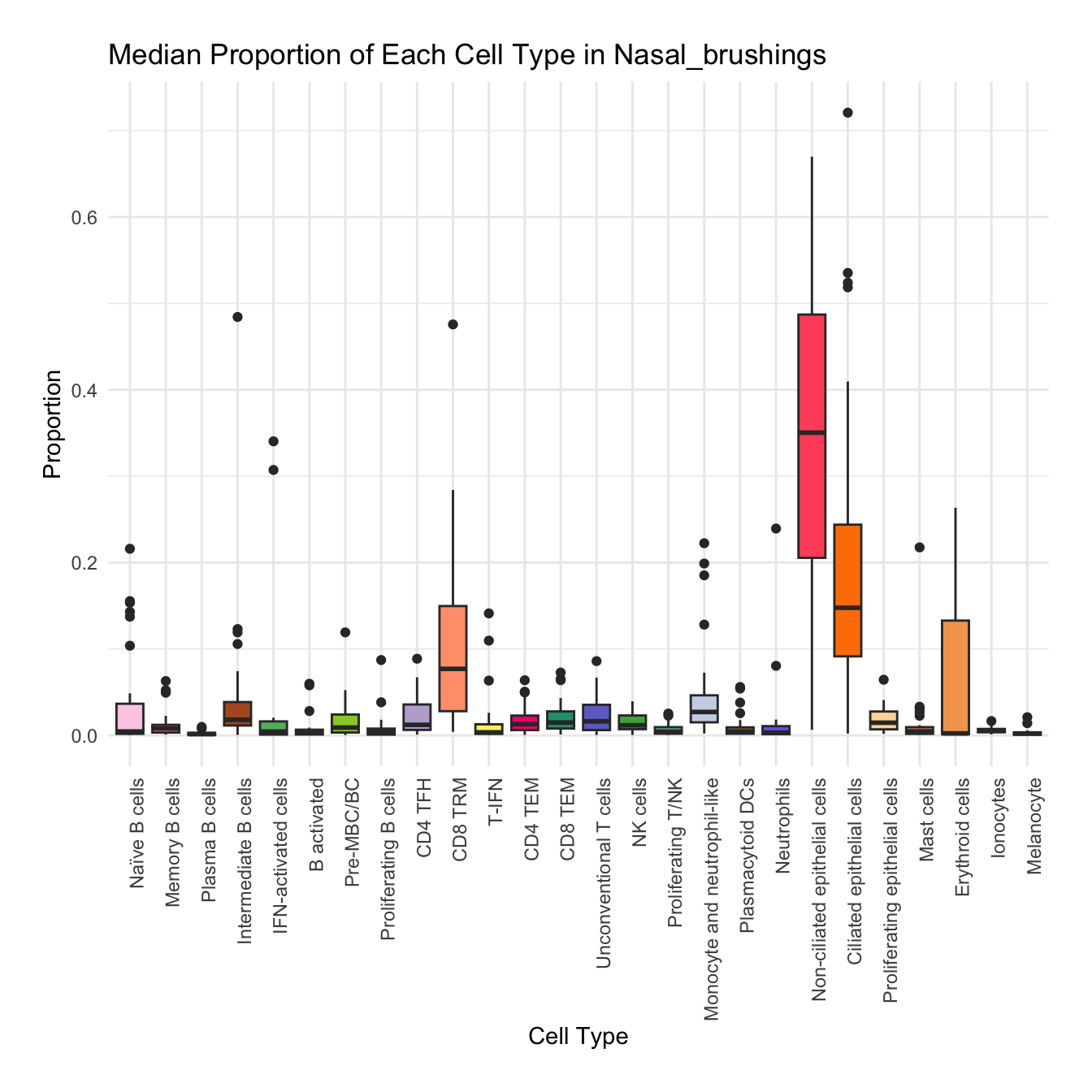
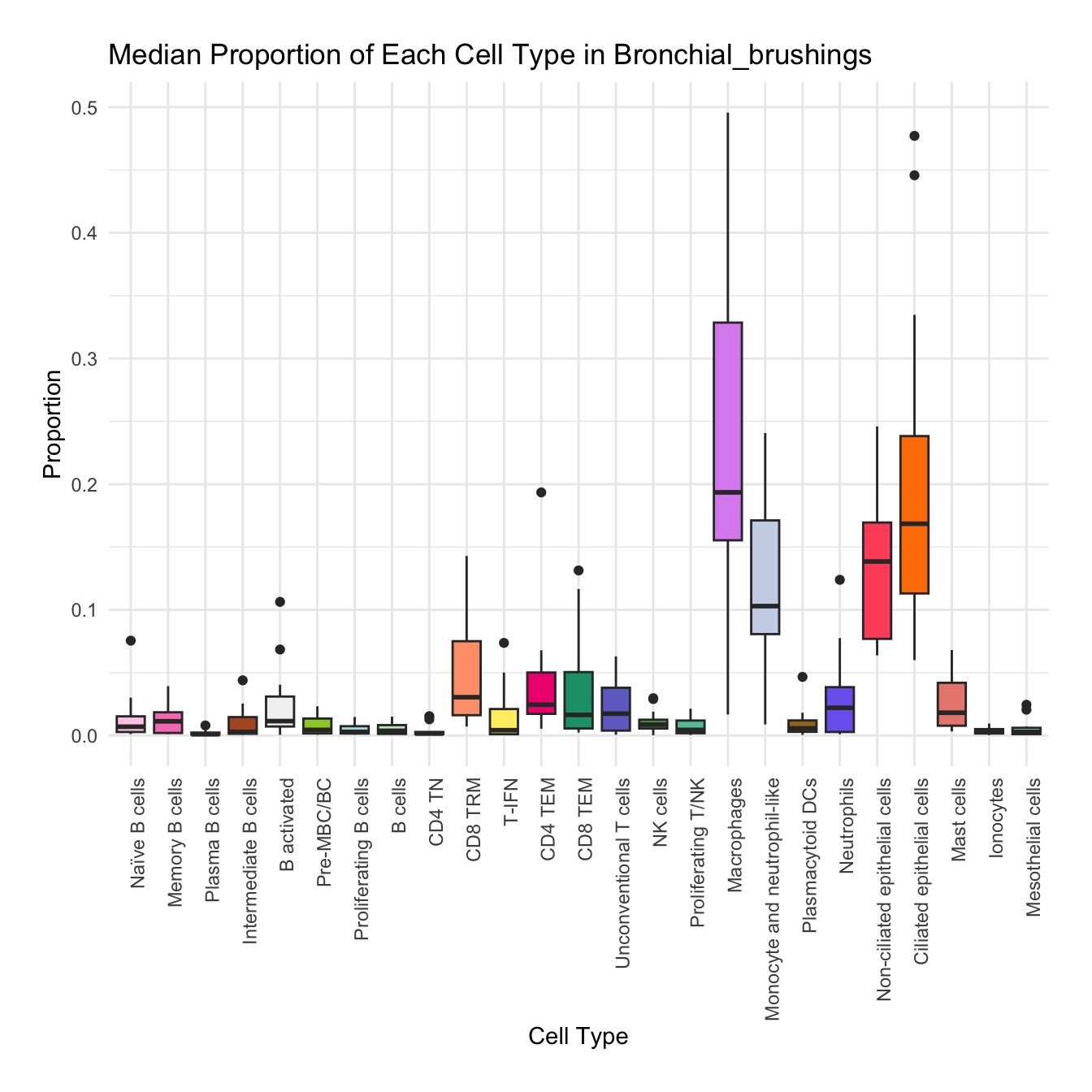
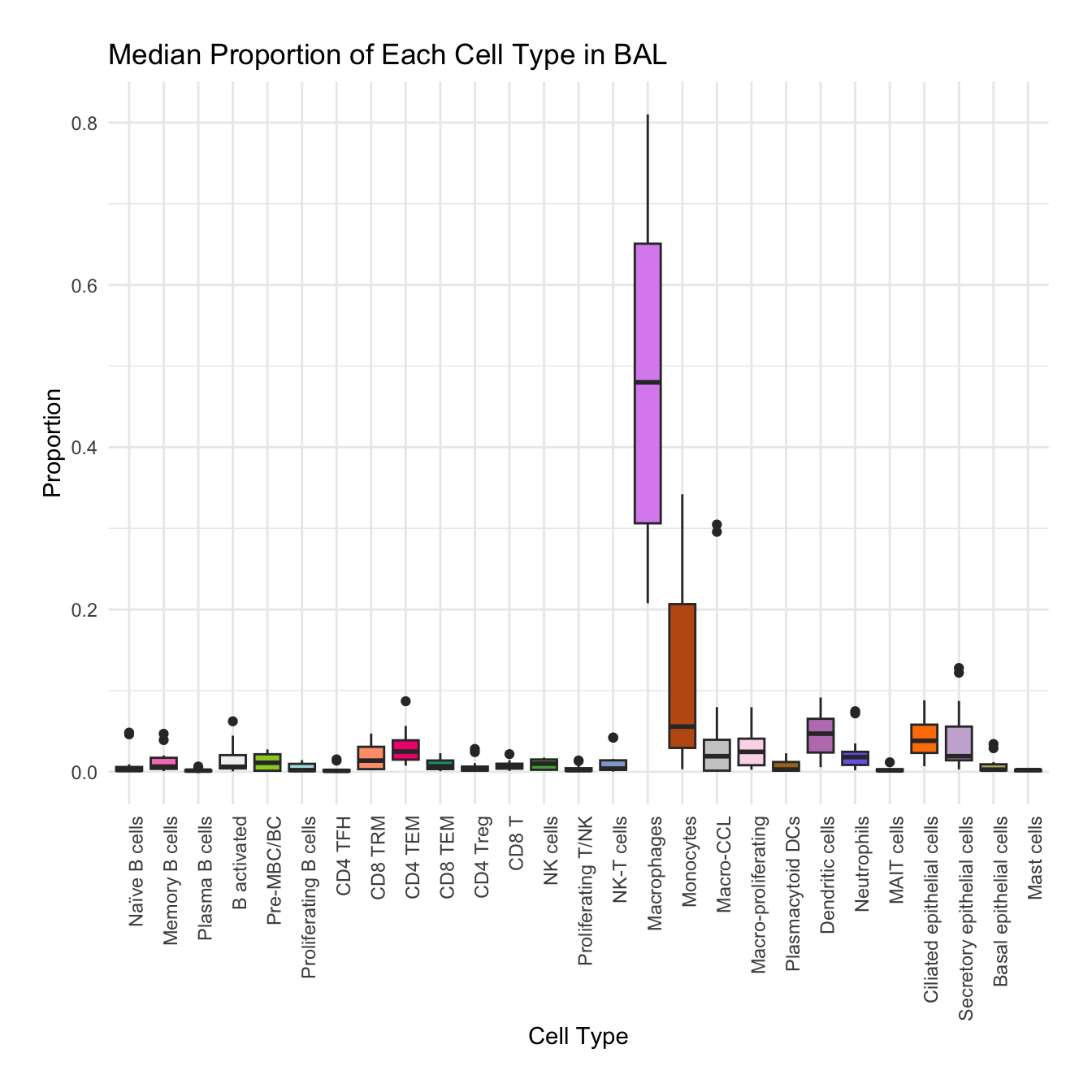
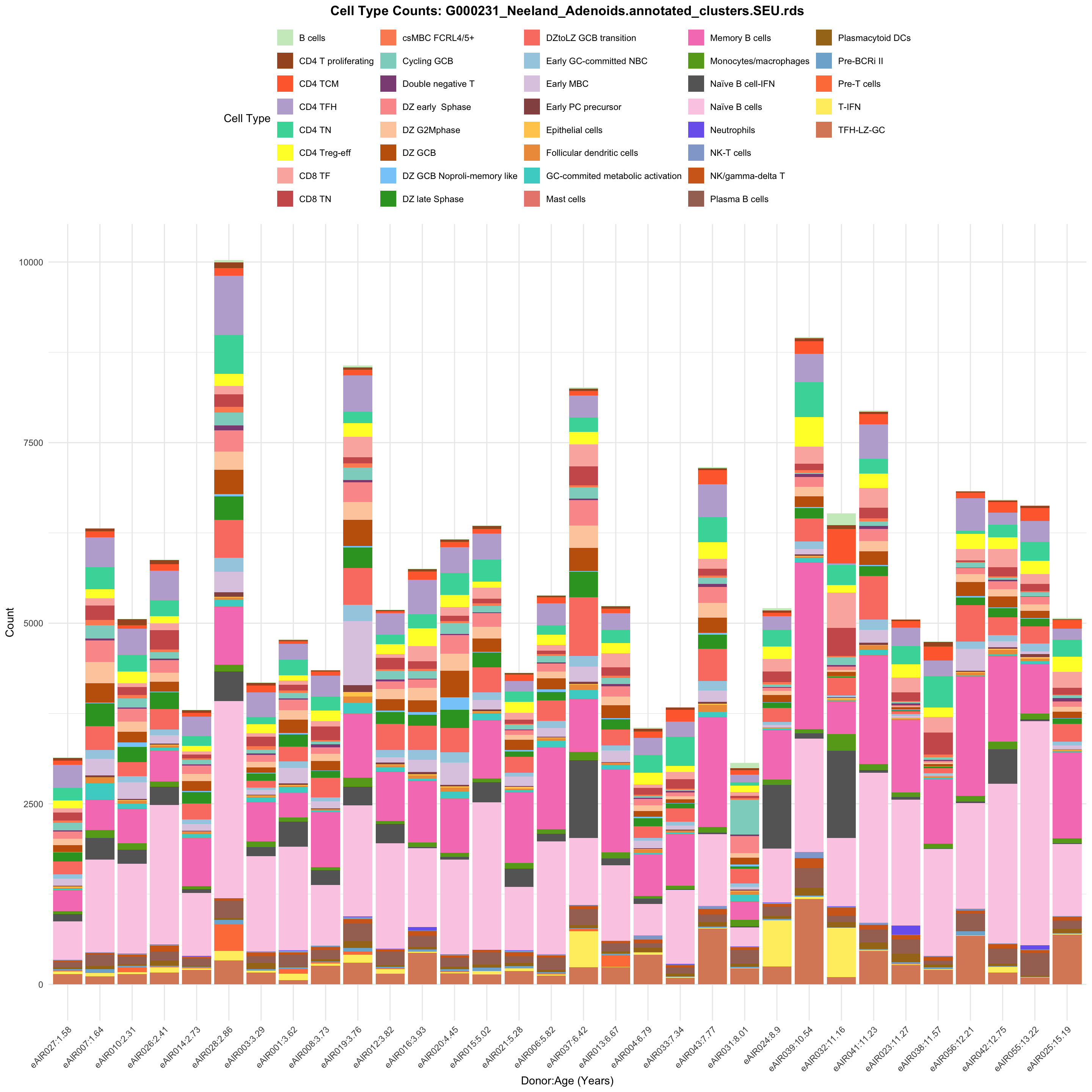
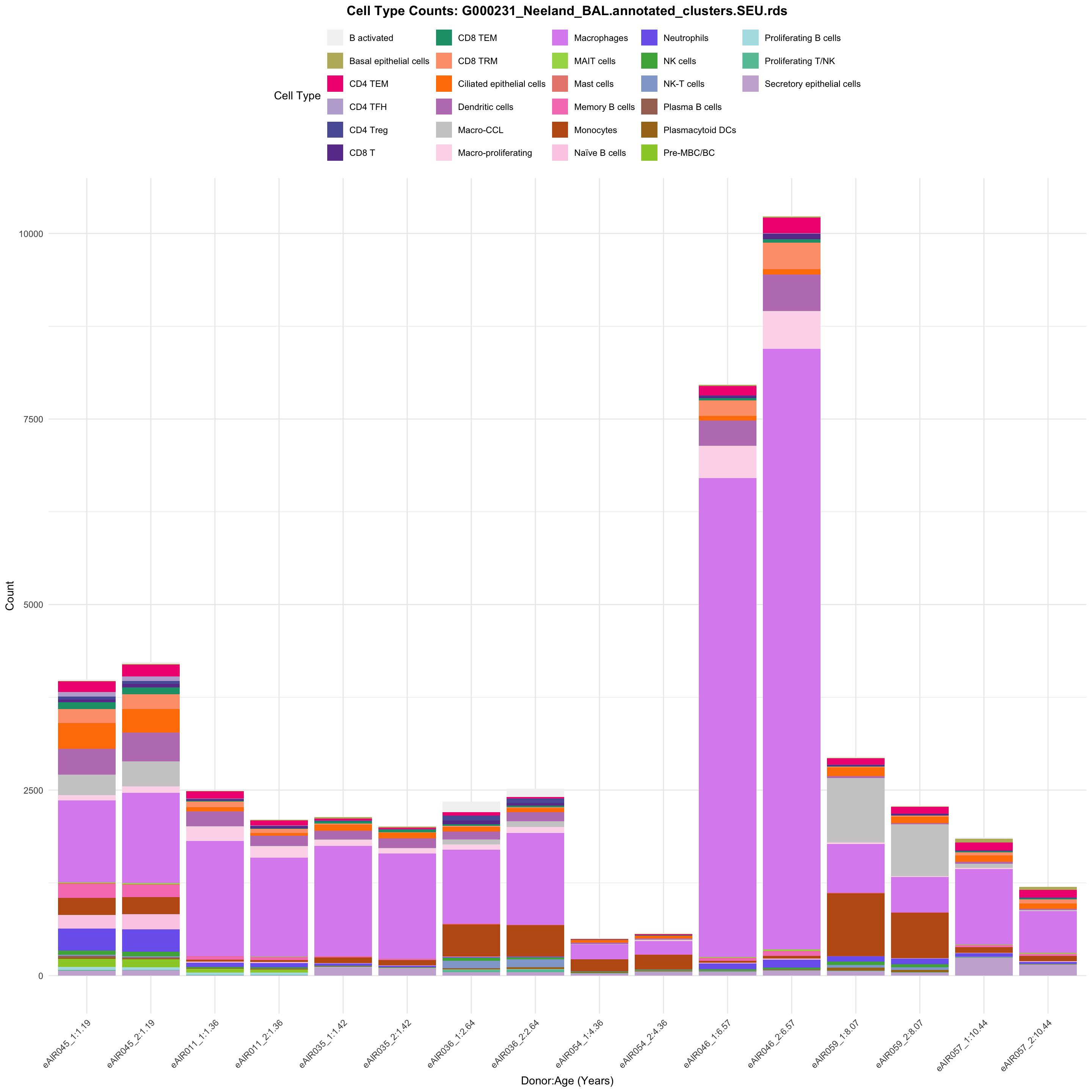
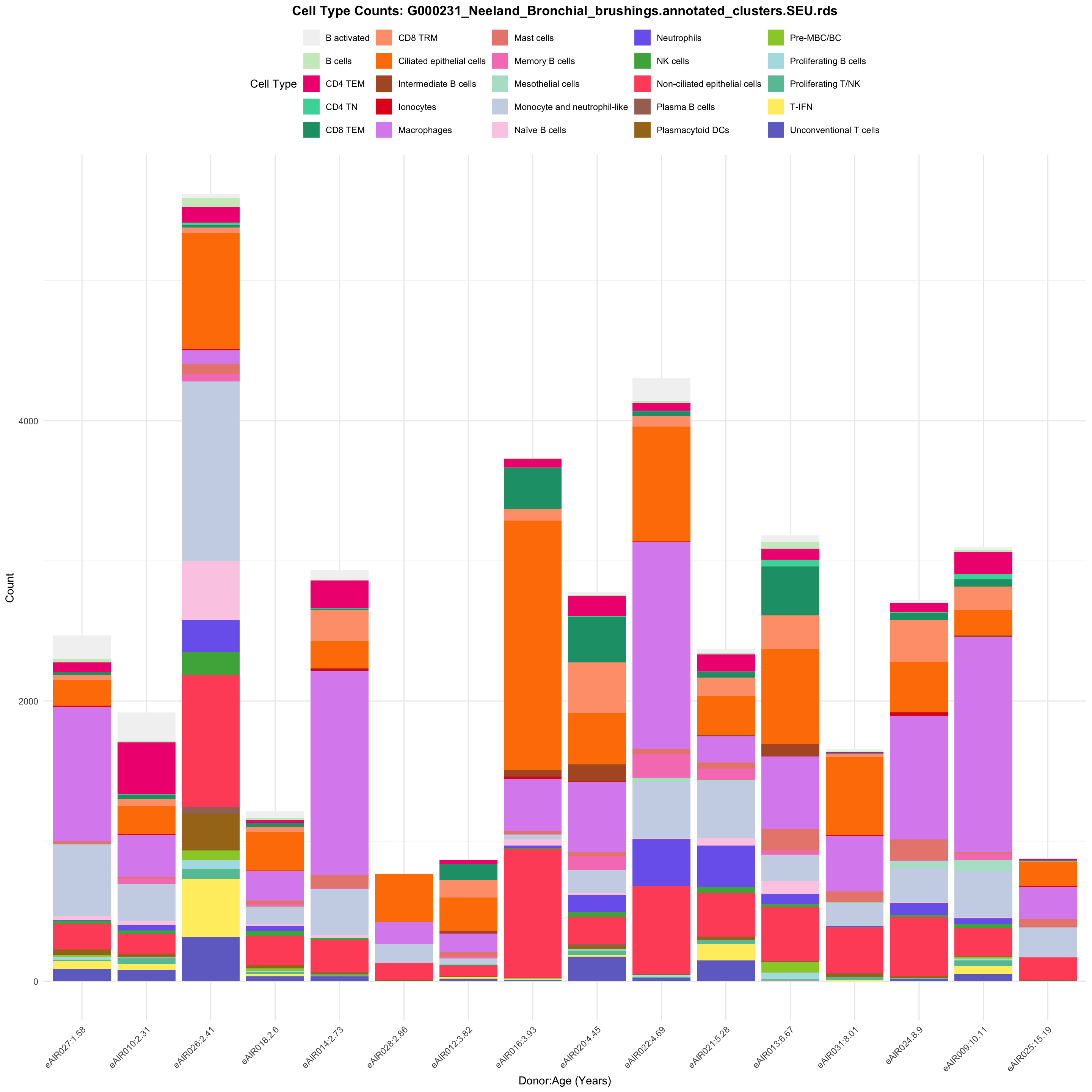
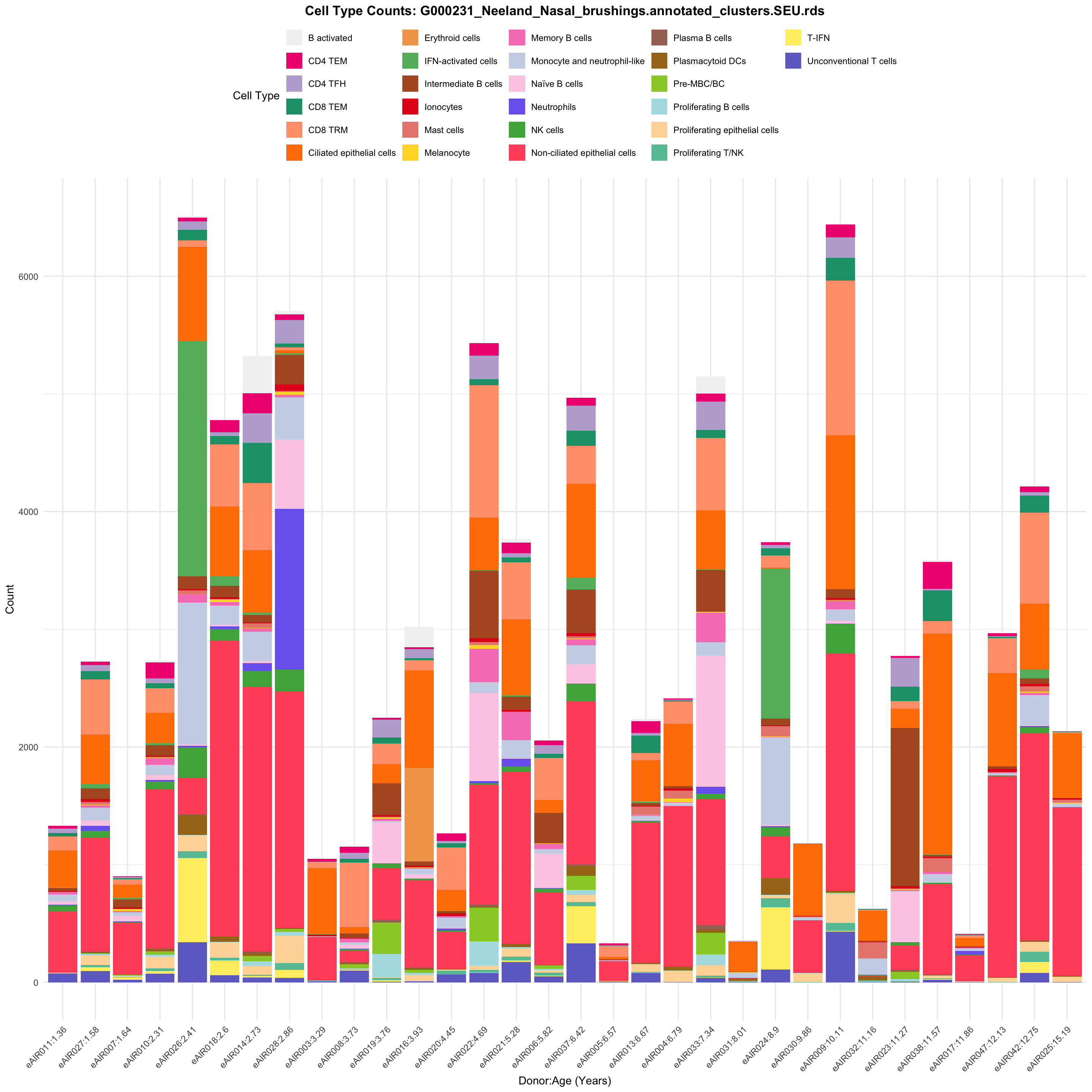
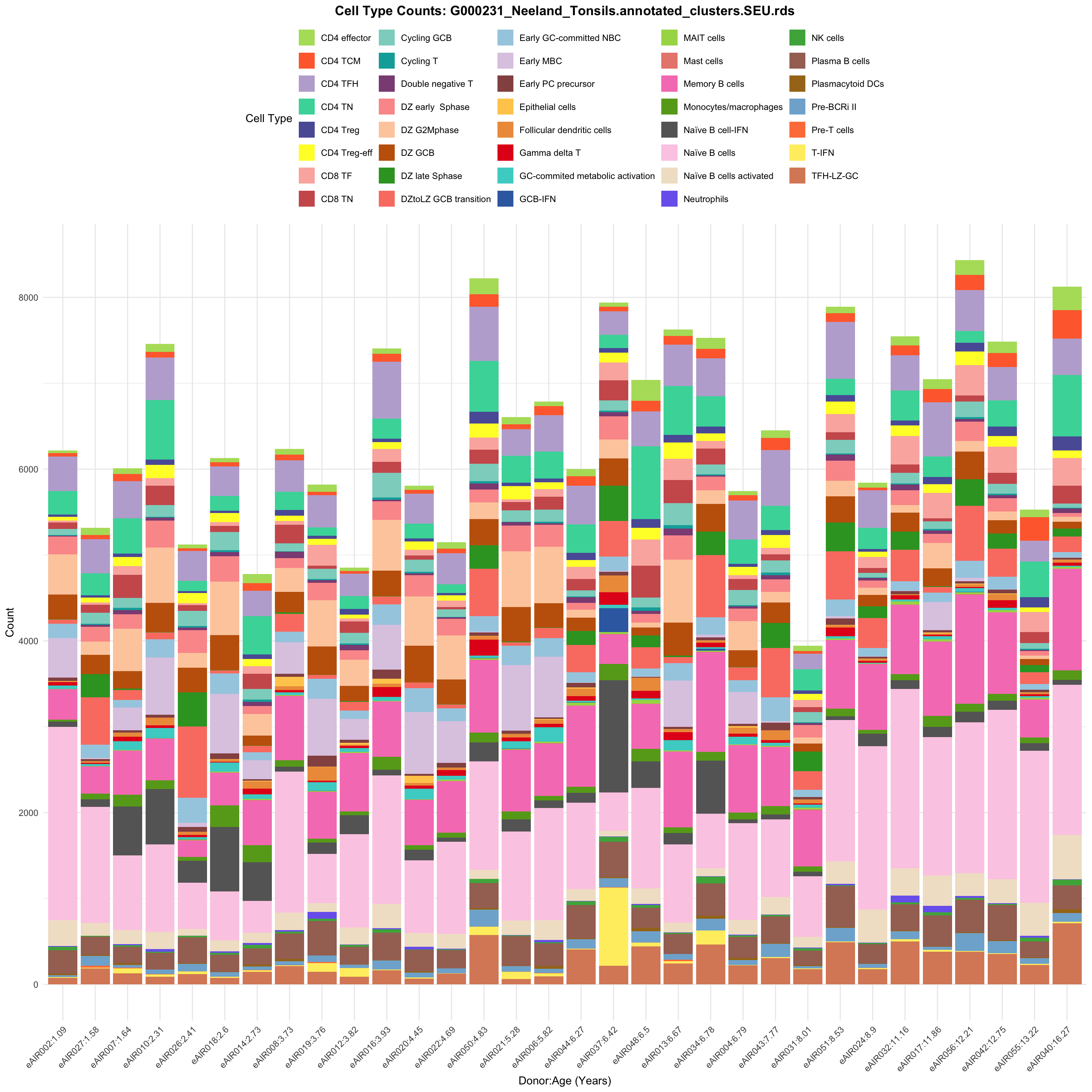
for (tissue in tissue_list) {
seu <- readRDS(tissue)
metadata_df <- data.frame(
sample = seu$sample_id,
donor = seu$donor_id,
age_years = as.character(seu$age_years),
cell_type = seu$cell_labels_v2
)
metadata_df$age_years <- as.numeric(metadata_df$age_years)
barplot_data <- metadata_df %>%
group_by(donor, age_years, cell_type) %>%
summarise(n_cells = n(), .groups = 'drop') %>%
arrange(donor, age_years)
p <- ggplot(barplot_data, aes(x = reorder(paste(donor, age_years, sep = ":"), age_years),
y = n_cells, fill = cell_type)) +
geom_bar(stat = "identity") +
ggtitle(paste0("Cell Type Counts: ", basename(tissue))) +
labs(x = "Donor:Age (Years)", y = "Count", fill = "Cell Type") +
scale_fill_manual(values = color_palette) +
theme_minimal() +
theme(
plot.title = element_text(size = 13, hjust = 0.5, face = "bold"),
legend.position = "top",
axis.text.x = element_text(angle = 45, hjust = 1)
)
print(p)
}
Session Info
sessionInfo()R version 4.3.2 (2023-10-31)
Platform: aarch64-apple-darwin20 (64-bit)
Running under: macOS 15.3
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: Australia/Melbourne
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] circlize_0.4.16 BiocParallel_1.36.0 harmony_1.2.0
[4] Rcpp_1.0.12 patchwork_1.2.0 cowplot_1.1.3
[7] data.table_1.15.0 RColorBrewer_1.1-3 kableExtra_1.4.0
[10] ggridges_0.5.6 ggforce_0.4.2 viridis_0.6.5
[13] viridisLite_0.4.2 paletteer_1.6.0 clustree_0.5.1
[16] ggraph_2.1.0 Seurat_5.0.1.9009 SeuratObject_5.0.1
[19] sp_2.1-3 here_1.0.1 lubridate_1.9.3
[22] forcats_1.0.0 stringr_1.5.1 dplyr_1.1.4
[25] purrr_1.0.2 readr_2.1.5 tidyr_1.3.1
[28] tibble_3.2.1 ggplot2_3.5.0 tidyverse_2.0.0
[31] BiocStyle_2.30.0 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] shape_1.4.6.1 rstudioapi_0.15.0 jsonlite_1.8.8
[4] magrittr_2.0.3 spatstat.utils_3.0-4 farver_2.1.1
[7] rmarkdown_2.25 GlobalOptions_0.1.2 fs_1.6.3
[10] vctrs_0.6.5 ROCR_1.0-11 spatstat.explore_3.2-6
[13] htmltools_0.5.7 sass_0.4.8 sctransform_0.4.1
[16] parallelly_1.37.0 KernSmooth_2.23-22 bslib_0.6.1
[19] htmlwidgets_1.6.4 ica_1.0-3 plyr_1.8.9
[22] plotly_4.10.4 zoo_1.8-12 cachem_1.0.8
[25] whisker_0.4.1 igraph_2.0.2 mime_0.12
[28] lifecycle_1.0.4 pkgconfig_2.0.3 Matrix_1.6-5
[31] R6_2.5.1 fastmap_1.1.1 fitdistrplus_1.1-11
[34] future_1.33.1 shiny_1.8.0 digest_0.6.34
[37] colorspace_2.1-0 rematch2_2.1.2 ps_1.7.6
[40] rprojroot_2.0.4 tensor_1.5 RSpectra_0.16-1
[43] irlba_2.3.5.1 labeling_0.4.3 progressr_0.14.0
[46] fansi_1.0.6 spatstat.sparse_3.0-3 timechange_0.3.0
[49] polyclip_1.10-6 httr_1.4.7 abind_1.4-5
[52] compiler_4.3.2 withr_3.0.0 fastDummies_1.7.3
[55] highr_0.10 MASS_7.3-60.0.1 tools_4.3.2
[58] lmtest_0.9-40 httpuv_1.6.14 future.apply_1.11.1
[61] goftest_1.2-3 glue_1.7.0 callr_3.7.5
[64] nlme_3.1-164 promises_1.2.1 grid_4.3.2
[67] Rtsne_0.17 getPass_0.2-4 cluster_2.1.6
[70] reshape2_1.4.4 generics_0.1.3 gtable_0.3.4
[73] spatstat.data_3.0-4 tzdb_0.4.0 hms_1.1.3
[76] xml2_1.3.6 tidygraph_1.3.1 utf8_1.2.4
[79] spatstat.geom_3.2-8 RcppAnnoy_0.0.22 ggrepel_0.9.5
[82] RANN_2.6.1 pillar_1.9.0 spam_2.10-0
[85] RcppHNSW_0.6.0 later_1.3.2 splines_4.3.2
[88] tweenr_2.0.3 lattice_0.22-5 deldir_2.0-2
[91] survival_3.5-8 tidyselect_1.2.0 miniUI_0.1.1.1
[94] pbapply_1.7-2 knitr_1.45 git2r_0.33.0
[97] gridExtra_2.3 svglite_2.1.3 scattermore_1.2
[100] xfun_0.42 graphlayouts_1.1.0 matrixStats_1.2.0
[103] stringi_1.8.3 lazyeval_0.2.2 yaml_2.3.8
[106] evaluate_0.23 codetools_0.2-19 BiocManager_1.30.22
[109] cli_3.6.2 uwot_0.1.16 systemfonts_1.0.5
[112] xtable_1.8-4 reticulate_1.35.0 munsell_0.5.0
[115] processx_3.8.3 jquerylib_0.1.4 spatstat.random_3.2-2
[118] globals_0.16.2 png_0.1-8 parallel_4.3.2
[121] ellipsis_0.3.2 dotCall64_1.1-1 listenv_0.9.1
[124] scales_1.3.0 leiden_0.4.3.1 rlang_1.1.3
sessionInfo()R version 4.3.2 (2023-10-31)
Platform: aarch64-apple-darwin20 (64-bit)
Running under: macOS 15.3
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: Australia/Melbourne
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] circlize_0.4.16 BiocParallel_1.36.0 harmony_1.2.0
[4] Rcpp_1.0.12 patchwork_1.2.0 cowplot_1.1.3
[7] data.table_1.15.0 RColorBrewer_1.1-3 kableExtra_1.4.0
[10] ggridges_0.5.6 ggforce_0.4.2 viridis_0.6.5
[13] viridisLite_0.4.2 paletteer_1.6.0 clustree_0.5.1
[16] ggraph_2.1.0 Seurat_5.0.1.9009 SeuratObject_5.0.1
[19] sp_2.1-3 here_1.0.1 lubridate_1.9.3
[22] forcats_1.0.0 stringr_1.5.1 dplyr_1.1.4
[25] purrr_1.0.2 readr_2.1.5 tidyr_1.3.1
[28] tibble_3.2.1 ggplot2_3.5.0 tidyverse_2.0.0
[31] BiocStyle_2.30.0 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] shape_1.4.6.1 rstudioapi_0.15.0 jsonlite_1.8.8
[4] magrittr_2.0.3 spatstat.utils_3.0-4 farver_2.1.1
[7] rmarkdown_2.25 GlobalOptions_0.1.2 fs_1.6.3
[10] vctrs_0.6.5 ROCR_1.0-11 spatstat.explore_3.2-6
[13] htmltools_0.5.7 sass_0.4.8 sctransform_0.4.1
[16] parallelly_1.37.0 KernSmooth_2.23-22 bslib_0.6.1
[19] htmlwidgets_1.6.4 ica_1.0-3 plyr_1.8.9
[22] plotly_4.10.4 zoo_1.8-12 cachem_1.0.8
[25] whisker_0.4.1 igraph_2.0.2 mime_0.12
[28] lifecycle_1.0.4 pkgconfig_2.0.3 Matrix_1.6-5
[31] R6_2.5.1 fastmap_1.1.1 fitdistrplus_1.1-11
[34] future_1.33.1 shiny_1.8.0 digest_0.6.34
[37] colorspace_2.1-0 rematch2_2.1.2 ps_1.7.6
[40] rprojroot_2.0.4 tensor_1.5 RSpectra_0.16-1
[43] irlba_2.3.5.1 labeling_0.4.3 progressr_0.14.0
[46] fansi_1.0.6 spatstat.sparse_3.0-3 timechange_0.3.0
[49] polyclip_1.10-6 httr_1.4.7 abind_1.4-5
[52] compiler_4.3.2 withr_3.0.0 fastDummies_1.7.3
[55] highr_0.10 MASS_7.3-60.0.1 tools_4.3.2
[58] lmtest_0.9-40 httpuv_1.6.14 future.apply_1.11.1
[61] goftest_1.2-3 glue_1.7.0 callr_3.7.5
[64] nlme_3.1-164 promises_1.2.1 grid_4.3.2
[67] Rtsne_0.17 getPass_0.2-4 cluster_2.1.6
[70] reshape2_1.4.4 generics_0.1.3 gtable_0.3.4
[73] spatstat.data_3.0-4 tzdb_0.4.0 hms_1.1.3
[76] xml2_1.3.6 tidygraph_1.3.1 utf8_1.2.4
[79] spatstat.geom_3.2-8 RcppAnnoy_0.0.22 ggrepel_0.9.5
[82] RANN_2.6.1 pillar_1.9.0 spam_2.10-0
[85] RcppHNSW_0.6.0 later_1.3.2 splines_4.3.2
[88] tweenr_2.0.3 lattice_0.22-5 deldir_2.0-2
[91] survival_3.5-8 tidyselect_1.2.0 miniUI_0.1.1.1
[94] pbapply_1.7-2 knitr_1.45 git2r_0.33.0
[97] gridExtra_2.3 svglite_2.1.3 scattermore_1.2
[100] xfun_0.42 graphlayouts_1.1.0 matrixStats_1.2.0
[103] stringi_1.8.3 lazyeval_0.2.2 yaml_2.3.8
[106] evaluate_0.23 codetools_0.2-19 BiocManager_1.30.22
[109] cli_3.6.2 uwot_0.1.16 systemfonts_1.0.5
[112] xtable_1.8-4 reticulate_1.35.0 munsell_0.5.0
[115] processx_3.8.3 jquerylib_0.1.4 spatstat.random_3.2-2
[118] globals_0.16.2 png_0.1-8 parallel_4.3.2
[121] ellipsis_0.3.2 dotCall64_1.1-1 listenv_0.9.1
[124] scales_1.3.0 leiden_0.4.3.1 rlang_1.1.3