Subclustering: Bronchial_brushings
Unsupervised Clustering of Broad cell labels
Gunjan Dixit
September 26, 2024
Last updated: 2024-09-26
Checks: 6 1
Knit directory: paed-airway-allTissues/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of
the R Markdown file created these results, you’ll want to first commit
it to the Git repo. If you’re still working on the analysis, you can
ignore this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230811) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 2376778. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: data/.DS_Store
Ignored: data/RDS/
Ignored: output/.DS_Store
Ignored: output/CSV/.DS_Store
Ignored: output/G000231_Neeland_batch1/
Ignored: output/G000231_Neeland_batch2_1/
Ignored: output/G000231_Neeland_batch2_2/
Ignored: output/G000231_Neeland_batch3/
Ignored: output/G000231_Neeland_batch4/
Ignored: output/G000231_Neeland_batch5/
Ignored: output/G000231_Neeland_batch9_1/
Ignored: output/RDS/
Ignored: output/plots/
Untracked files:
Untracked: Adenoids_Bcell_subset_proportions_Age.pdf
Untracked: Adenoids_Tcell_subset_proportions_Age.pdf
Untracked: Adenoids_cell_type_proportions_Age.pdf
Untracked: Age_proportions_Adenoids.pdf
Untracked: Age_proportions_Bronchial_brushings.pdf
Untracked: Age_proportions_Nasal_brushings.pdf
Untracked: Age_proportions_Tonsils.pdf
Untracked: BAL_Tcell_propeller.xlsx
Untracked: BAL_propeller.xlsx
Untracked: BB_Tcell_propeller.xlsx
Untracked: BB_propeller.xlsx
Untracked: NB_Tcell_propeller.xlsx
Untracked: NB_propeller.csv
Untracked: NB_propeller.pdf
Untracked: NB_propeller.xlsx
Untracked: Tonsils_cell_type_proportions.jpg
Untracked: Tonsils_cell_type_proportions.pdf
Untracked: Tonsils_cell_type_proportions.png
Untracked: Tonsils_cell_type_proportions_Age.pdf
Untracked: analysis/03_Batch_Integration.Rmd
Untracked: analysis/Age_modelling_Adenoids.Rmd
Untracked: analysis/Age_modelling_Bronchial_Brushings.Rmd
Untracked: analysis/Age_modelling_Nasal_Brushings.Rmd
Untracked: analysis/Age_modelling_Tonsils.Rmd
Untracked: analysis/Age_proportions.Rmd
Untracked: analysis/Age_proportions_AllBatches.Rmd
Untracked: analysis/BAL_without_DecontX.Rmd
Untracked: analysis/Batch_Integration_&_Downstream_analysis.Rmd
Untracked: analysis/Batch_correction_&_Downstream.Rmd
Untracked: analysis/Boxplot_Adenoids.pdf
Untracked: analysis/Boxplot_BAL.pdf
Untracked: analysis/Boxplot_Bronchial_brushings.pdf
Untracked: analysis/Boxplot_Nasal_brushings.pdf
Untracked: analysis/Boxplot_Tonsils.pdf
Untracked: analysis/Cell_cycle_regression.Rmd
Untracked: analysis/Master_metadata.Rmd
Untracked: analysis/Preprocessing_Batch1_Nasal_brushings.Rmd
Untracked: analysis/Preprocessing_Batch2_Tonsils.Rmd
Untracked: analysis/Preprocessing_Batch3_Adenoids.Rmd
Untracked: analysis/Preprocessing_Batch4_Bronchial_brushings.Rmd
Untracked: analysis/Preprocessing_Batch5_Nasal_brushings.Rmd
Untracked: analysis/Preprocessing_Batch6_BAL.Rmd
Untracked: analysis/Preprocessing_Batch7_Bronchial_brushings.Rmd
Untracked: analysis/Preprocessing_Batch8_Adenoids.Rmd
Untracked: analysis/Preprocessing_Batch9_Tonsils.Rmd
Untracked: analysis/TonsilsVsAdenoids.Rmd
Untracked: analysis/boxplot_proportions_Adenoids.pdf
Untracked: analysis/boxplot_proportions_BAL.pdf
Untracked: analysis/boxplot_proportions_Bronchial_brushings.pdf
Untracked: analysis/boxplot_proportions_Nasal_brushings.pdf
Untracked: analysis/boxplot_proportions_Tonsils.pdf
Untracked: analysis/boxplot_proportions__broad_l2Adenoids.pdf
Untracked: analysis/boxplot_proportions__broad_l2BAL.pdf
Untracked: analysis/boxplot_proportions__broad_l2Bronchial_brushings.pdf
Untracked: analysis/boxplot_proportions__broad_l2Nasal_brushings.pdf
Untracked: analysis/boxplot_proportions__broad_l2Tonsils.pdf
Untracked: analysis/cell_cycle_regression.R
Untracked: analysis/test.Rmd
Untracked: analysis/testing_age_all.Rmd
Untracked: cell_proportions_overview.png
Untracked: cell_type_proportions.pdf
Untracked: cell_type_proportions_enhanced.pdf
Untracked: cell_type_proportions_individual.pdf
Untracked: color_palette.rds
Untracked: color_palette_v2_level2.rds
Untracked: combined_metadata.rds
Untracked: data/Cell_labels_Mel/
Untracked: data/Cell_labels_Mel_v2/
Untracked: data/Cell_labels_modified_Gunjan/
Untracked: data/Hs.c2.cp.reactome.v7.1.entrez.rds
Untracked: data/Raw_feature_bc_matrix/
Untracked: data/celltypes_Mel_GD_v3.xlsx
Untracked: data/celltypes_Mel_GD_v4_no_dups.xlsx
Untracked: data/celltypes_Mel_modified.xlsx
Untracked: data/celltypes_Mel_v2.csv
Untracked: data/celltypes_Mel_v2.xlsx
Untracked: data/celltypes_Mel_v2_MN.xlsx
Untracked: data/celltypes_for_mel_MN.xlsx
Untracked: data/earlyAIR_sample_sheets_combined.xlsx
Untracked: output/CSV/All_tissues.propeller.xlsx
Untracked: output/CSV/Bronchial_brushings/
Untracked: output/CSV/Bronchial_brushings_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/
Untracked: output/CSV/Bronchial_brushings_Marker_genes_Reclustered_Bcell_population.RNA_snn_res.0.3/
Untracked: output/CSV/G000231_Neeland_Adenoids.propeller.xlsx
Untracked: output/CSV/G000231_Neeland_Bronchial_brushings.propeller.xlsx
Untracked: output/CSV/G000231_Neeland_Nasal_brushings.propeller.xlsx
Untracked: output/CSV/G000231_Neeland_Tonsils.propeller.xlsx
Untracked: output/CSV/Nasal_brushings/
Unstaged changes:
Deleted: 02_QC_exploratoryPlots.Rmd
Deleted: 02_QC_exploratoryPlots.html
Modified: analysis/00_AllBatches_overview.Rmd
Modified: analysis/01_QC_emptyDrops.Rmd
Modified: analysis/02_QC_exploratoryPlots.Rmd
Modified: analysis/Adenoids.Rmd
Modified: analysis/Age_modeling.Rmd
Modified: analysis/AllBatches_QCExploratory.Rmd
Modified: analysis/BAL.Rmd
Modified: analysis/Bronchial_brushings.Rmd
Modified: analysis/Nasal_brushings.Rmd
Modified: analysis/Subclustering_Bronchial_brushings.Rmd
Modified: analysis/Tonsils.Rmd
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c0.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c1.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c10.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c11.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c12.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c13.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c14.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c15.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c16.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c17.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c2.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c3.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c4.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c5.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c6.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c7.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c8.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/REACTOME-cluster-limma-c9.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c0.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c1.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c10.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c11.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c12.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c13.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c14.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c15.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c16.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c17.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c2.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c3.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c4.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c5.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c6.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c7.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c8.csv
Modified: output/CSV/BAL_Marker_gene_clusters.limmaTrendRNA_snn_res.0.4/up-cluster-limma-c9.csv
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown
(analysis/Subclustering_Bronchial_brushings.Rmd) and HTML
(docs/Subclustering_Bronchial_brushings.html) files. If
you’ve configured a remote Git repository (see
?wflow_git_remote), click on the hyperlinks in the table
below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 07af966 | Gunjan Dixit | 2024-09-25 | Modified index |
| Rmd | 3b5ab22 | Gunjan Dixit | 2024-09-25 | Separated Subclustering Rmd |
Introduction
Load libraries
suppressPackageStartupMessages({
library(BiocStyle)
library(tidyverse)
library(here)
library(glue)
library(dplyr)
library(Seurat)
library(clustree)
library(kableExtra)
library(RColorBrewer)
library(data.table)
library(ggplot2)
library(patchwork)
library(limma)
library(edgeR)
library(speckle)
library(AnnotationDbi)
library(org.Hs.eg.db)
library(readxl)
})Load Input data
For Bronchial brushings, we used only Batch4 for the downstream analysis.
tissue <- "Bronchial_brushings"
out1 <- here("output",
"RDS", "AllBatches_Clustering_SEUs",
paste0("G000231_Neeland_",tissue,".Clusters.SEU.rds"))
seu_obj <- readRDS(out1)
seu_objAn object of class Seurat
18046 features across 33917 samples within 1 assay
Active assay: RNA (18046 features, 2000 variable features)
3 layers present: counts, data, scale.data
3 dimensional reductions calculated: pca, umap, umap.unintegratedReclustering T cell population
This includes CD4 T cell, CD8 T cell, NK cell, NK-T cell, proliferating or cycling T/NK cell.
The marker genes for this reclustering can be found here-
idx <- which(Idents(seu_obj) %in% c("CD4 T cells", "CD8 T cells", "proliferating T/NK"))
paed_sub <- seu_obj[,idx]
mito_genes <- grep("^MT-", rownames(paed_sub), value = TRUE)
paed_sub <- subset(paed_sub, features = setdiff(rownames(paed_sub), mito_genes))
paed_subAn object of class Seurat
18035 features across 7632 samples within 1 assay
Active assay: RNA (18035 features, 1995 variable features)
3 layers present: counts, data, scale.data
3 dimensional reductions calculated: pca, umap, umap.unintegratedpaed_sub <- paed_sub %>%
NormalizeData() %>%
FindVariableFeatures() %>%
ScaleData() %>%
RunPCA()
paed_sub <- RunUMAP(paed_sub, dims = 1:30, reduction = "pca", reduction.name = "umap.new")
meta_data <- colnames(paed_sub@meta.data)
drop <- grep("^RNA_snn_res", meta_data, value = TRUE)
paed_sub@meta.data <- paed_sub@meta.data[, !(colnames(paed_sub@meta.data) %in% drop)]
resolutions <- seq(0.1, 1, by = 0.1)
paed_sub <- FindNeighbors(paed_sub, reduction = "pca", dims = 1:30)
paed_sub <- FindClusters(paed_sub, resolution = resolutions, algorithm = 3)Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 7632
Number of edges: 279916
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.9447
Number of communities: 7
Elapsed time: 4 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 7632
Number of edges: 279916
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.9200
Number of communities: 9
Elapsed time: 3 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 7632
Number of edges: 279916
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.9041
Number of communities: 10
Elapsed time: 3 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 7632
Number of edges: 279916
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.8890
Number of communities: 11
Elapsed time: 3 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 7632
Number of edges: 279916
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.8743
Number of communities: 12
Elapsed time: 3 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 7632
Number of edges: 279916
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.8605
Number of communities: 13
Elapsed time: 2 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 7632
Number of edges: 279916
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.8484
Number of communities: 15
Elapsed time: 2 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 7632
Number of edges: 279916
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.8392
Number of communities: 18
Elapsed time: 2 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 7632
Number of edges: 279916
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.8308
Number of communities: 20
Elapsed time: 2 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 7632
Number of edges: 279916
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.8230
Number of communities: 21
Elapsed time: 2 secondsDimHeatmap(paed_sub, dims = 1:10, cells = 500, balanced = TRUE)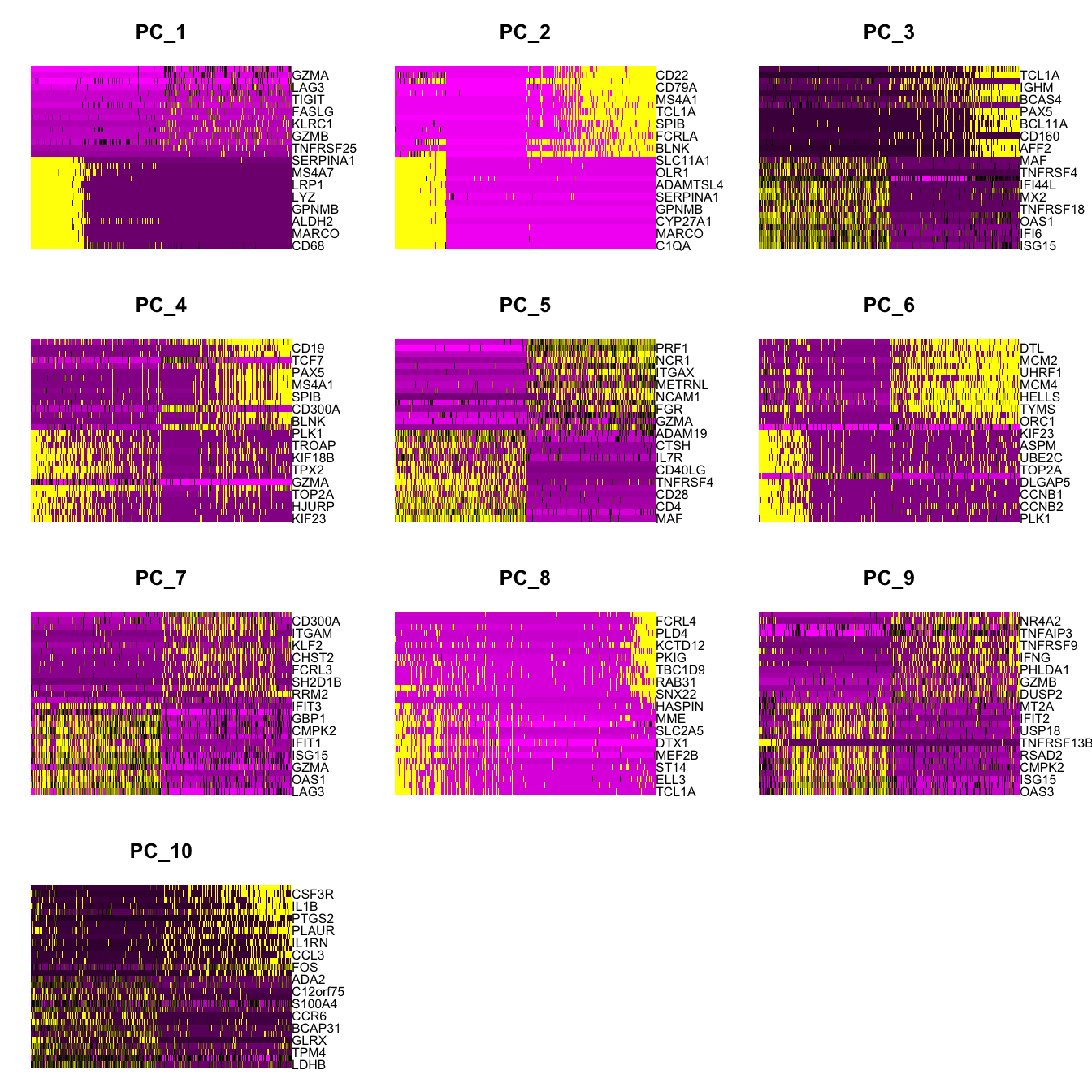
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
clustree(paed_sub, prefix = "RNA_snn_res.")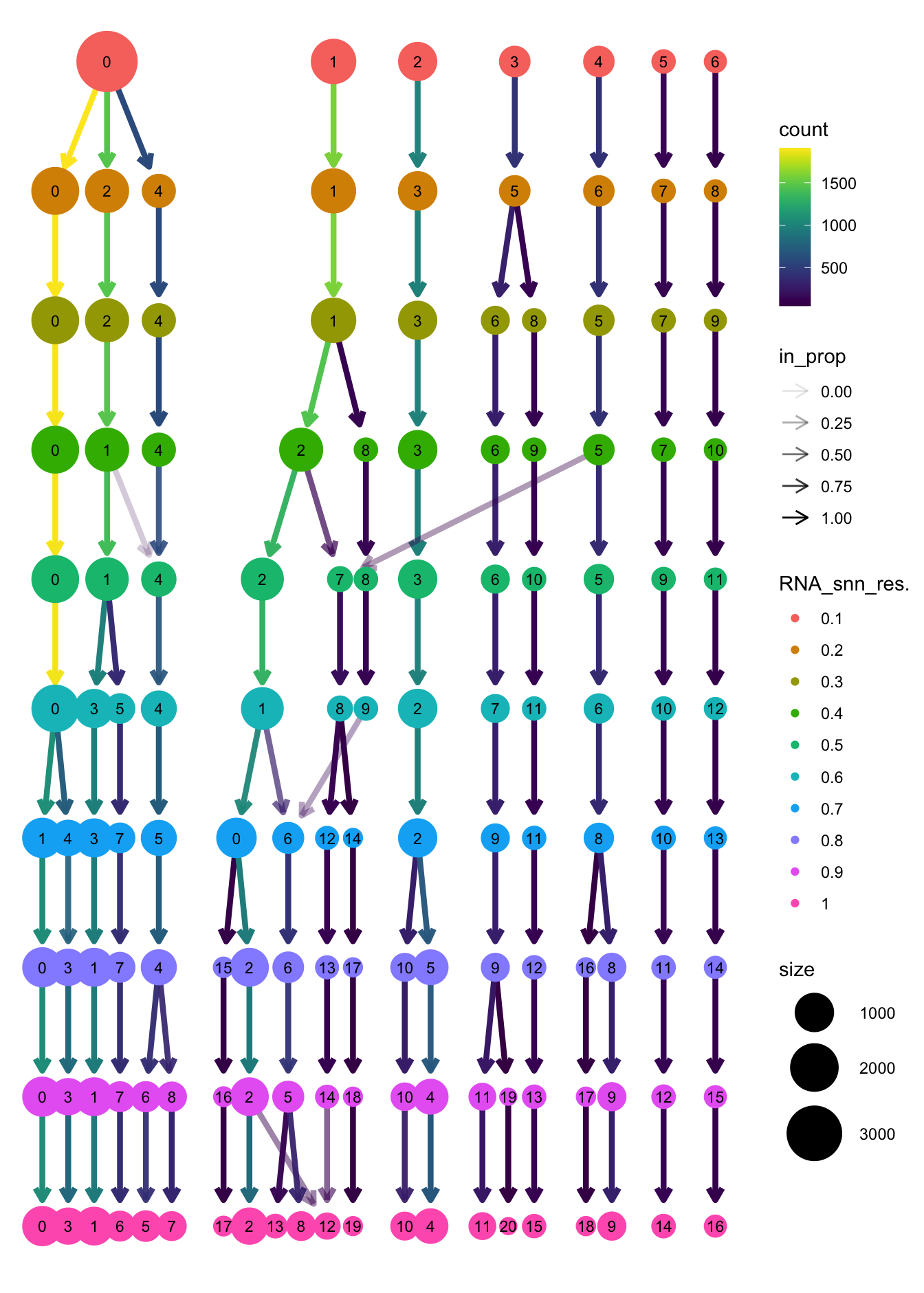
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
opt_res <- "RNA_snn_res.0.4"
n <- nlevels(paed_sub$RNA_snn_res.0.4)
paed_sub$RNA_snn_res.0.4 <- factor(paed_sub$RNA_snn_res.0.4, levels = seq(0,n-1))
paed_sub$seurat_clusters <- NULL
paed_sub$cluster <- paed_sub$RNA_snn_res.0.4
Idents(paed_sub) <- paed_sub$clusterpaed_sub.markers <- FindAllMarkers(paed_sub, only.pos = TRUE, min.pct = 0.25, logfc.threshold = 0.25)Calculating cluster 0Calculating cluster 1Calculating cluster 2Calculating cluster 3Calculating cluster 4Calculating cluster 5Calculating cluster 6Calculating cluster 7Calculating cluster 8Calculating cluster 9Calculating cluster 10paed_sub.markers %>%
group_by(cluster) %>% unique() %>%
top_n(n = 5, wt = avg_log2FC) -> top5
paed_sub.markers %>%
group_by(cluster) %>%
slice_head(n=1) %>%
pull(gene) -> best.wilcox.gene.per.cluster
best.wilcox.gene.per.cluster [1] "CD8A" "HOPX" "MAF" "TCF7" "LAG3" "TYROBP" "UHRF1"
[8] "CSF3R" "NOG" "POU2AF1" "GPNMB" Feature plot shows the expression of top marker genes per cluster.
FeaturePlot(paed_sub,features=best.wilcox.gene.per.cluster, reduction = 'umap.new', raster = FALSE, ncol = 2, label = TRUE)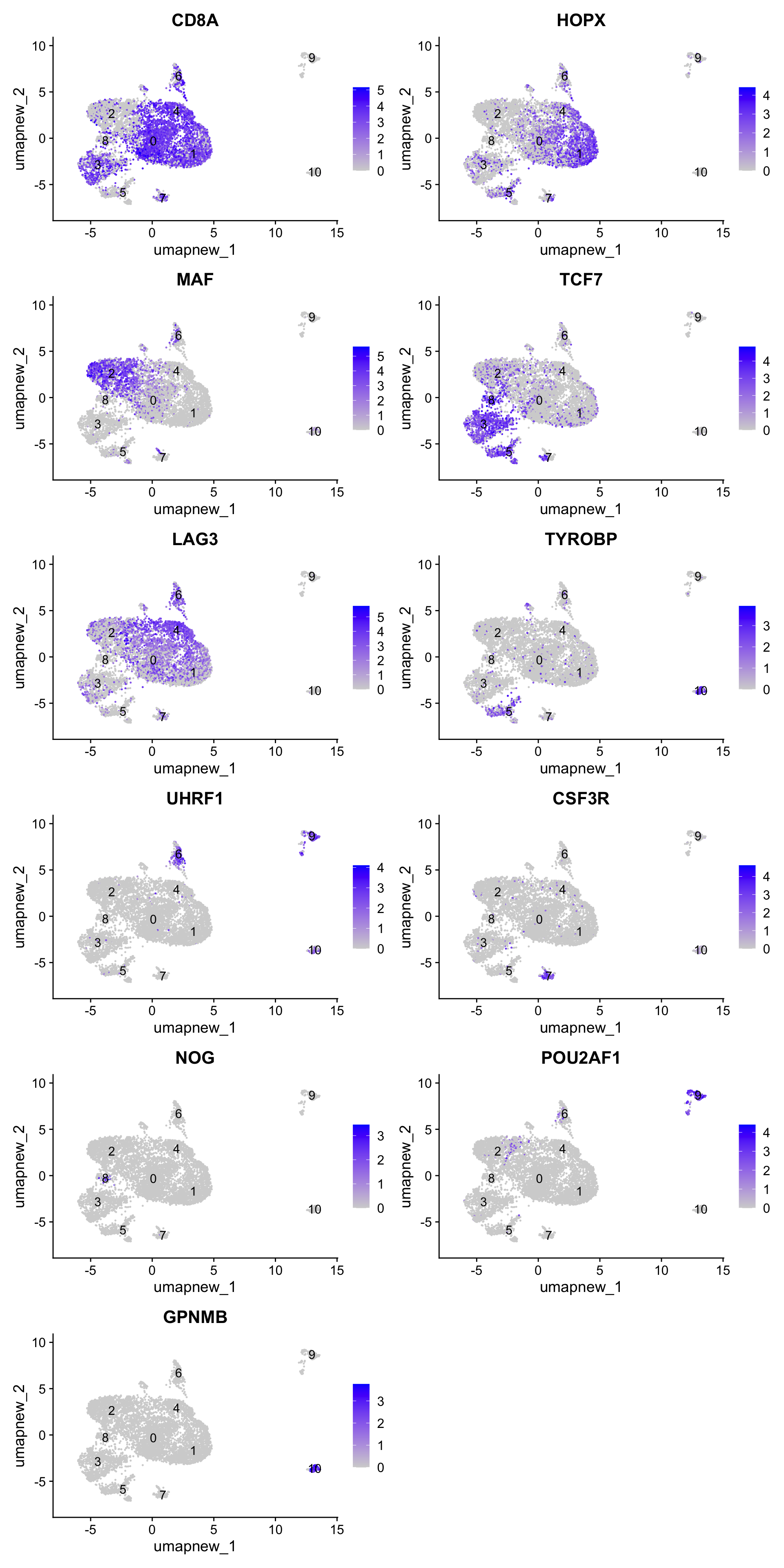
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
Top 10 marker genes from Seurat
## Seurat top markers
top10 <- paed_sub.markers %>%
group_by(cluster) %>%
top_n(n = 10, wt = avg_log2FC) %>%
ungroup() %>%
distinct(gene, .keep_all = TRUE) %>%
arrange(cluster, desc(avg_log2FC))
cluster_colors <- paletteer::paletteer_d("pals::glasbey")[factor(top10$cluster)]
DotPlot(paed_sub,
features = unique(top10$gene),
group.by = opt_res,
cols = c("azure1", "blueviolet"),
dot.scale = 3, assay = "RNA") +
RotatedAxis() +
FontSize(y.text = 8, x.text = 12) +
labs(y = element_blank(), x = element_blank()) +
coord_flip() +
theme(axis.text.y = element_text(color = cluster_colors)) +
ggtitle("Top 10 marker genes per cluster (Seurat)")Warning: Vectorized input to `element_text()` is not officially supported.
ℹ Results may be unexpected or may change in future versions of ggplot2.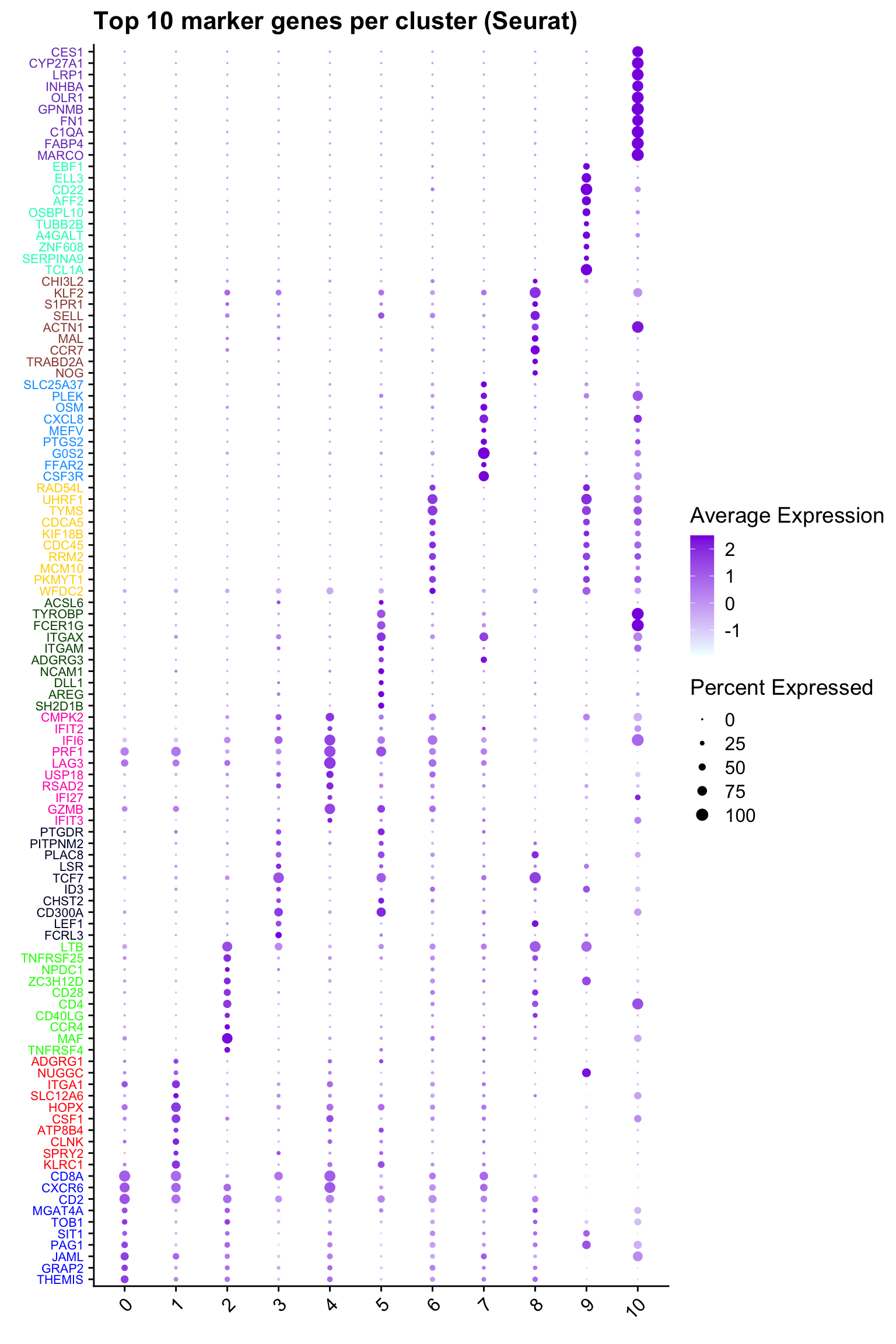
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
out_markers <- here("output",
"CSV",
paste(tissue,"_Marker_genes_Reclustered_Tcell_population.",opt_res, sep = ""))
dir.create(out_markers, recursive = TRUE, showWarnings = FALSE)
for (cl in unique(paed_sub.markers$cluster)) {
cluster_data <- paed_sub.markers %>% dplyr::filter(cluster == cl)
file_name <- here(out_markers, paste0("G000231_Neeland_",tissue, "_cluster_", cl, ".csv"))
if (!file.exists(file_name)) {
write.csv(cluster_data, file = file_name)
}
}Update T cell subclustering labels
cell_labels <- readxl::read_excel(here("data/Cell_labels_Mel_v2/earlyAIR_NB_BB_BAL_T-NK_annotations_16.07.24.xlsx"), sheet = "BB")
new_cluster_names <- cell_labels %>%
dplyr::select(cluster, annotation) %>%
deframe()
paed_sub <- RenameIdents(paed_sub, new_cluster_names)
paed_sub@meta.data$cell_labels_v2 <- Idents(paed_sub)
DimPlot(paed_sub, reduction = "umap.new", raster = FALSE, repel = TRUE, label = TRUE, label.size = 3.5) + ggtitle(paste0(tissue, ": UMAP with Updated subclustering"))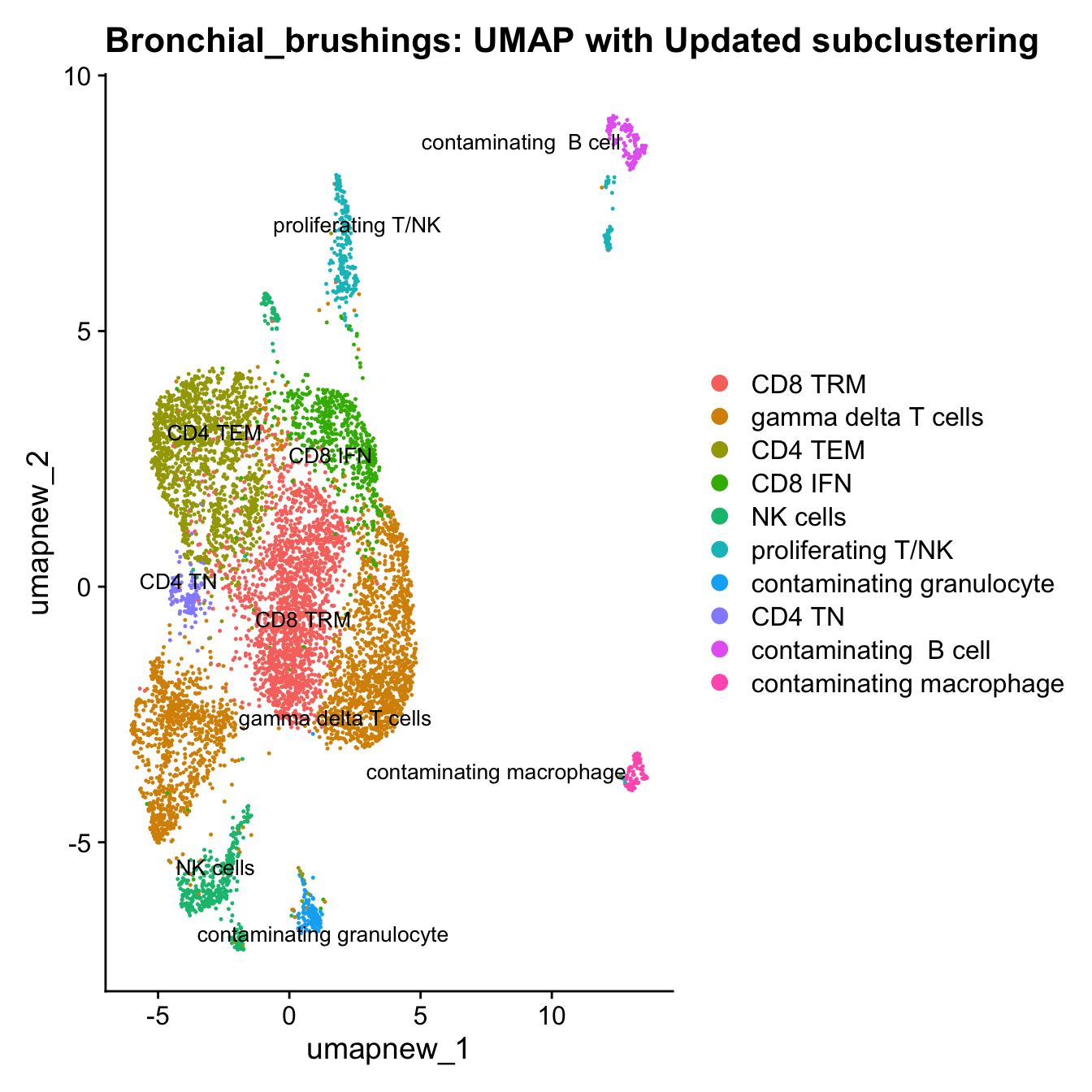
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
Summary Plots
palette1 <- paletteer::paletteer_d("ggthemes::Classic_20")
palette2 <- paletteer::paletteer_d("Polychrome::light")
combined_palette <- unique(c(palette1, palette2))
labels <- c( "predicted.ann_level_1","predicted.ann_level_2", "predicted.ann_level_3", "predicted.ann_level_4", "predicted.ann_level_5","predicted.ann_finest_level", "cell_labels", "cell_labels_v2", "RNA_snn_res.0.4")
p <- vector("list",length(labels))
for(label in labels){
paed_sub@meta.data %>%
ggplot(aes(x = !!sym(label),
fill = !!sym(label))) +
geom_bar() +
geom_text(aes(label = ..count..), stat = "count",
vjust = -0.5, colour = "black", size = 2) +
scale_y_log10() +
theme(axis.text.x = element_blank(),
axis.title.x = element_blank(),
axis.ticks.x = element_blank()) +
NoLegend() +
labs(y = "No. Cells (log scale)") -> p1
paed_sub@meta.data %>%
dplyr::select(!!sym(label), Sample) %>%
group_by(!!sym(label), Sample) %>%
summarise(num = n()) %>%
mutate(prop = num / sum(num)) %>%
ggplot(aes(x = !!sym(label), y = prop * 100,
fill = Sample)) +
geom_bar(stat = "identity") +
theme(axis.text.x = element_text(angle = 90,
vjust = 0.5,
hjust = 1,
size = 8)) +
labs(y = "% Cells", fill = "Sample") +
scale_fill_manual(values = combined_palette) -> p2
(p1 / p2) & theme(legend.text = element_text(size = 8),
legend.key.size = unit(3, "mm")) -> p[[label]]
}`summarise()` has grouped output by 'predicted.ann_level_1'. You can override
using the `.groups` argument.
`summarise()` has grouped output by 'predicted.ann_level_2'. You can override
using the `.groups` argument.
`summarise()` has grouped output by 'predicted.ann_level_3'. You can override
using the `.groups` argument.
`summarise()` has grouped output by 'predicted.ann_level_4'. You can override
using the `.groups` argument.
`summarise()` has grouped output by 'predicted.ann_level_5'. You can override
using the `.groups` argument.
`summarise()` has grouped output by 'predicted.ann_finest_level'. You can
override using the `.groups` argument.
`summarise()` has grouped output by 'cell_labels'. You can override using the
`.groups` argument.
`summarise()` has grouped output by 'cell_labels_v2'. You can override using
the `.groups` argument.
`summarise()` has grouped output by 'RNA_snn_res.0.4'. You can override using
the `.groups` argument.p[[1]]
NULL
[[2]]
NULL
[[3]]
NULL
[[4]]
NULL
[[5]]
NULL
[[6]]
NULL
[[7]]
NULL
[[8]]
NULL
[[9]]
NULL
$predicted.ann_level_1Warning: The dot-dot notation (`..count..`) was deprecated in ggplot2 3.4.0.
ℹ Please use `after_stat(count)` instead.
This warning is displayed once every 8 hours.
Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
generated.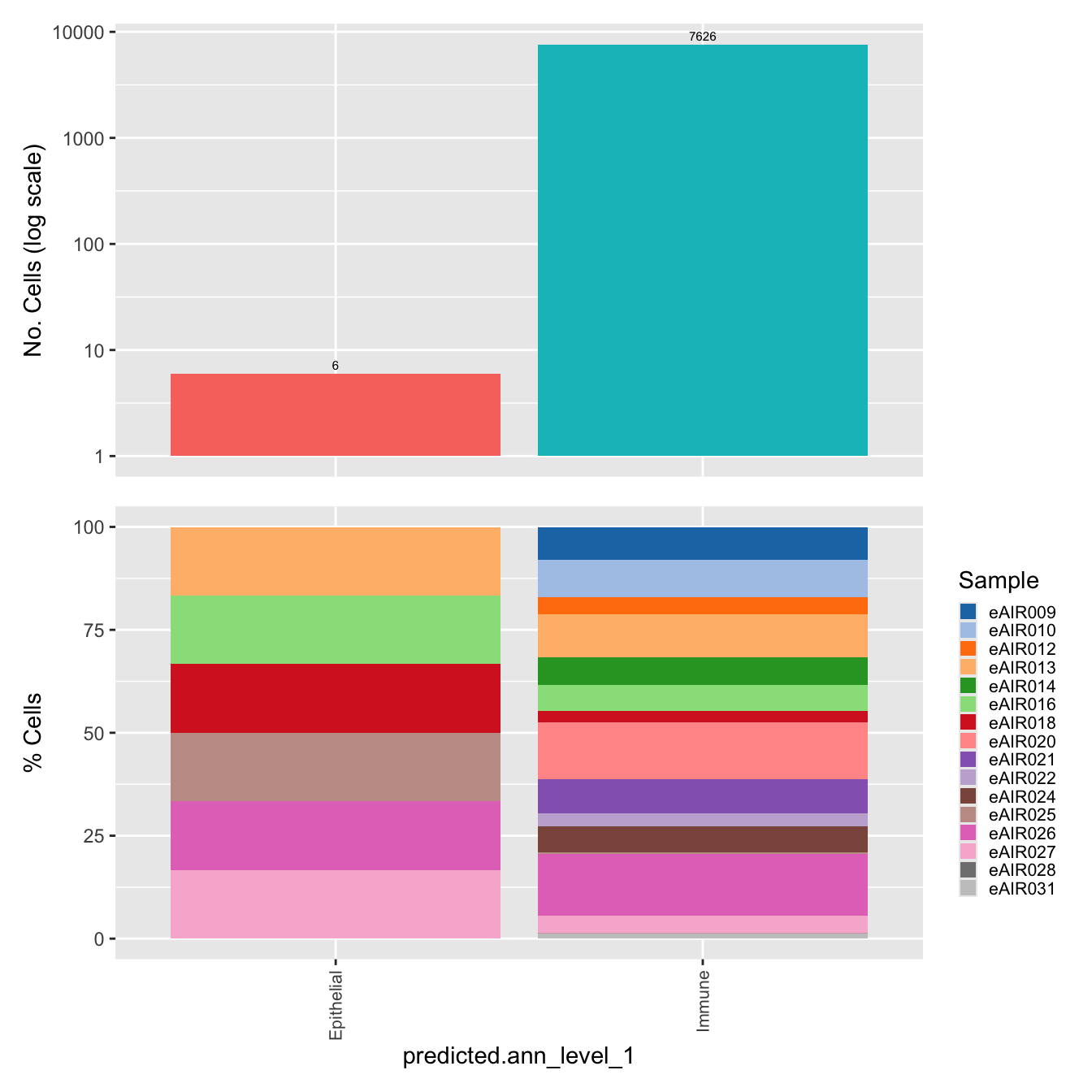
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
$predicted.ann_level_2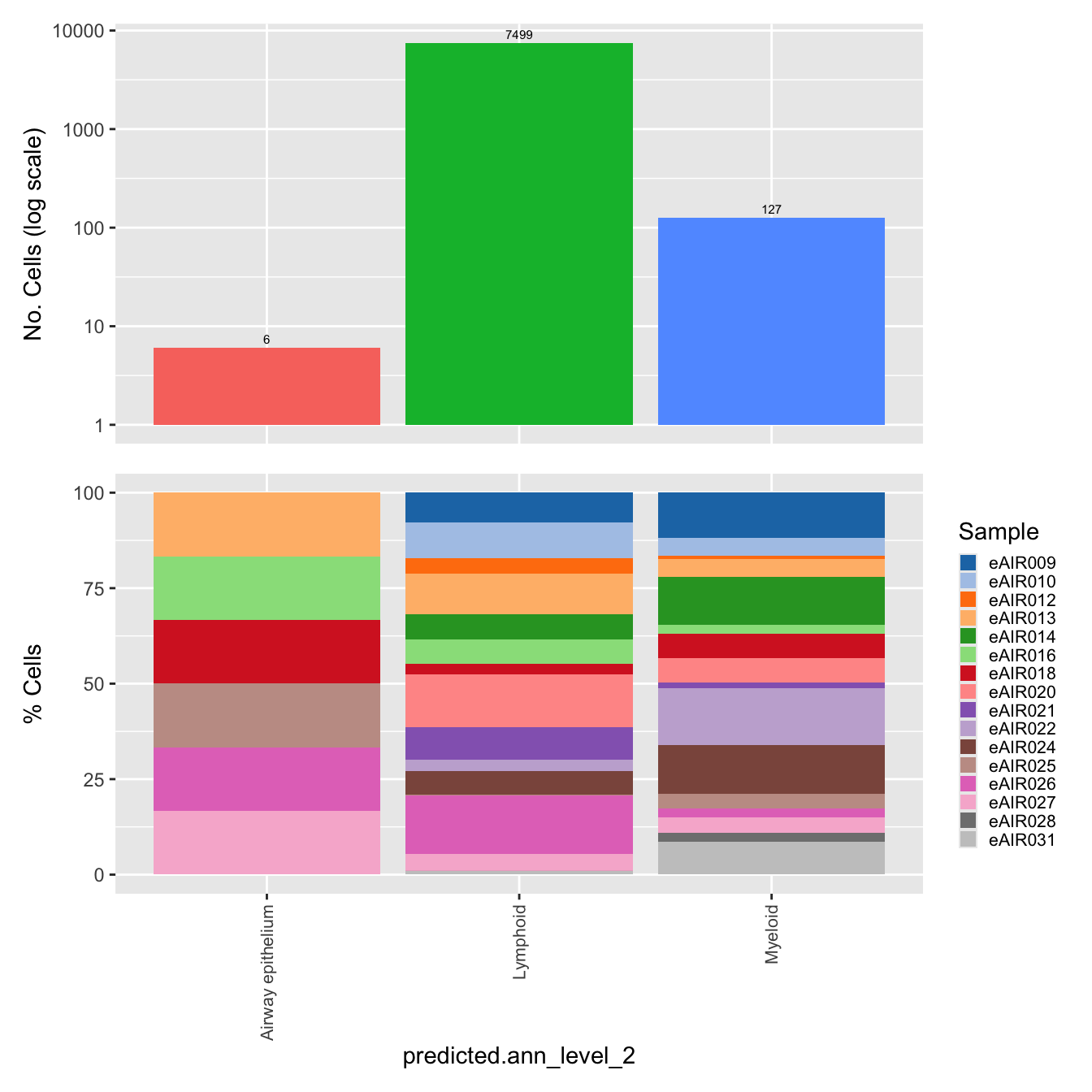
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
$predicted.ann_level_3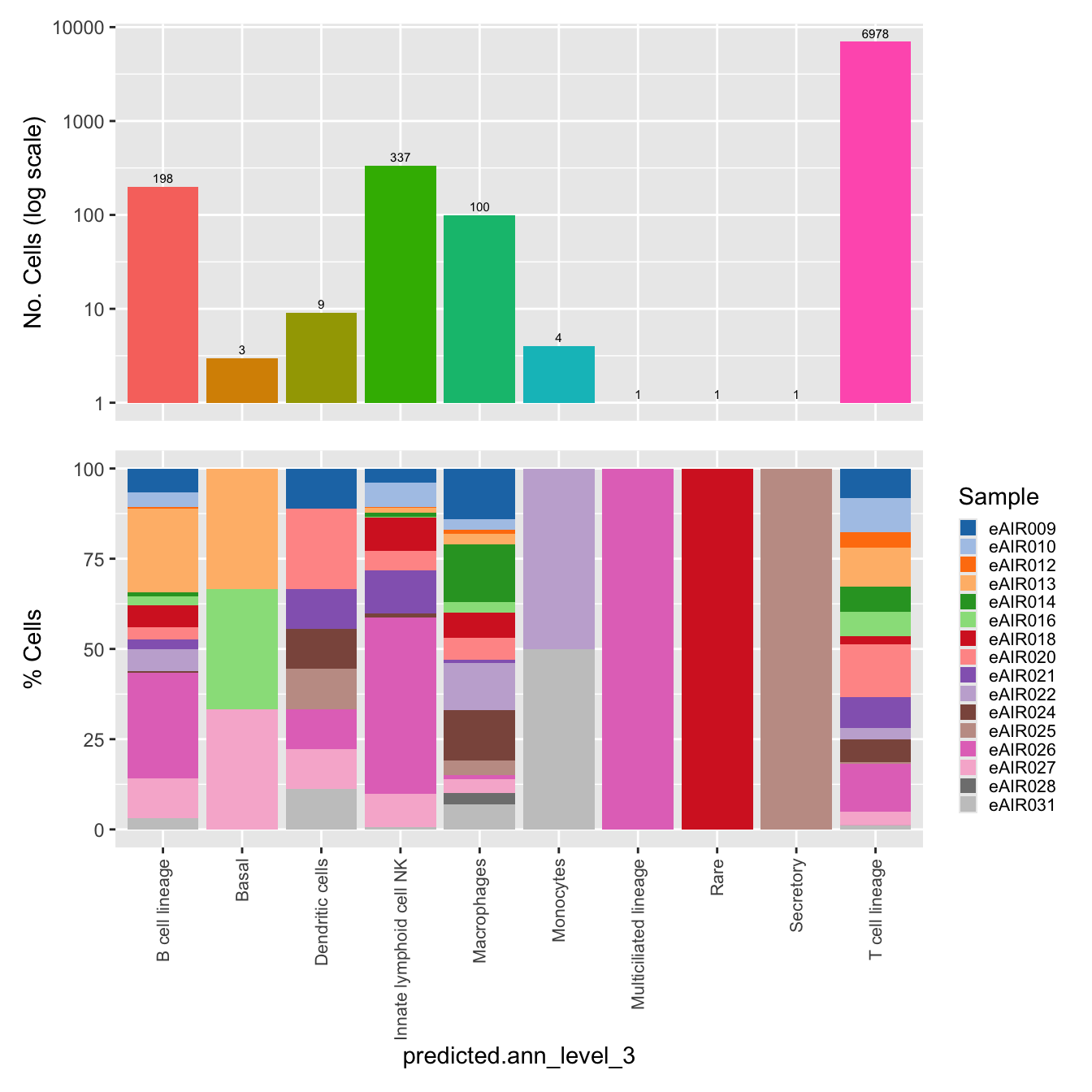
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
$predicted.ann_level_4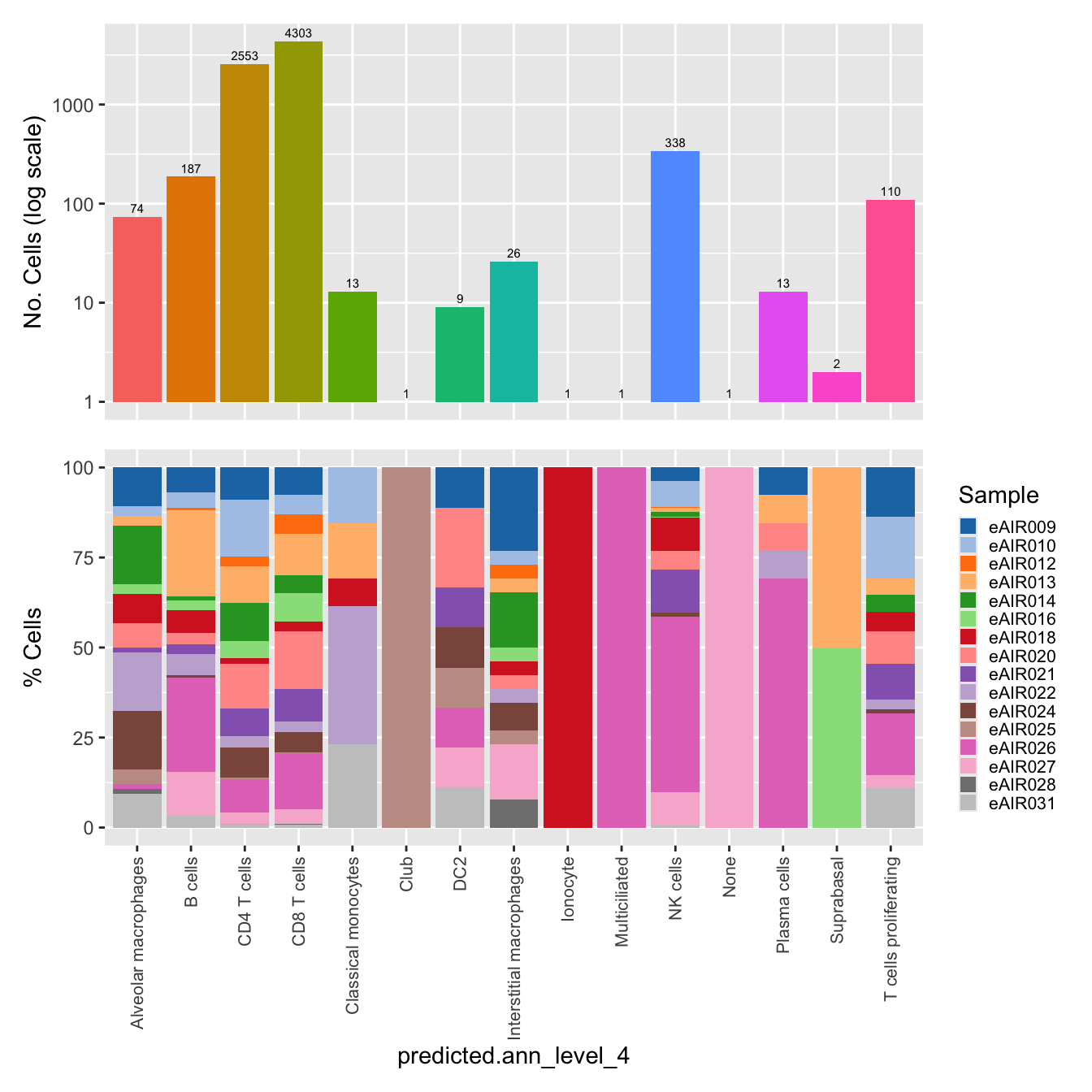
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
$predicted.ann_level_5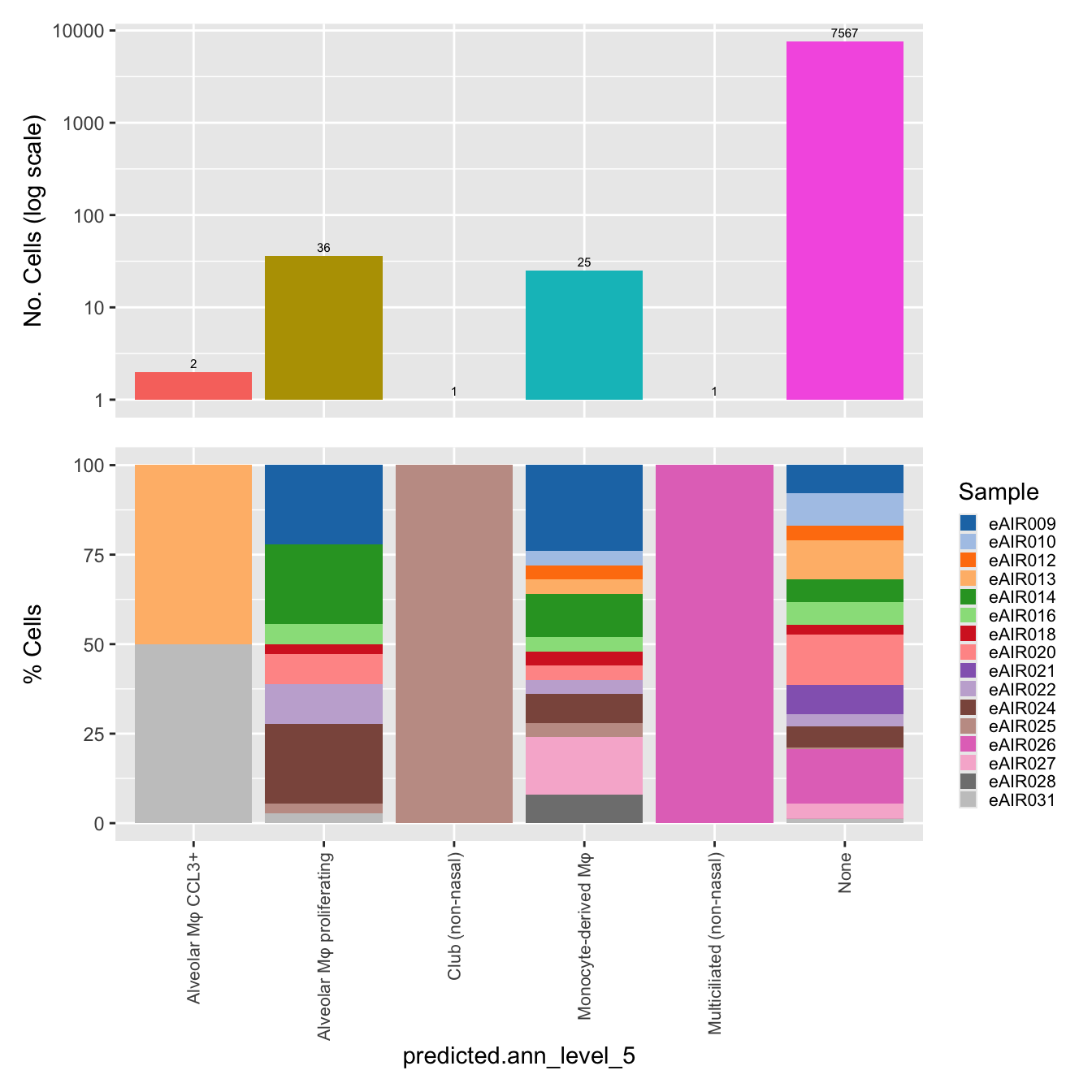
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
$predicted.ann_finest_level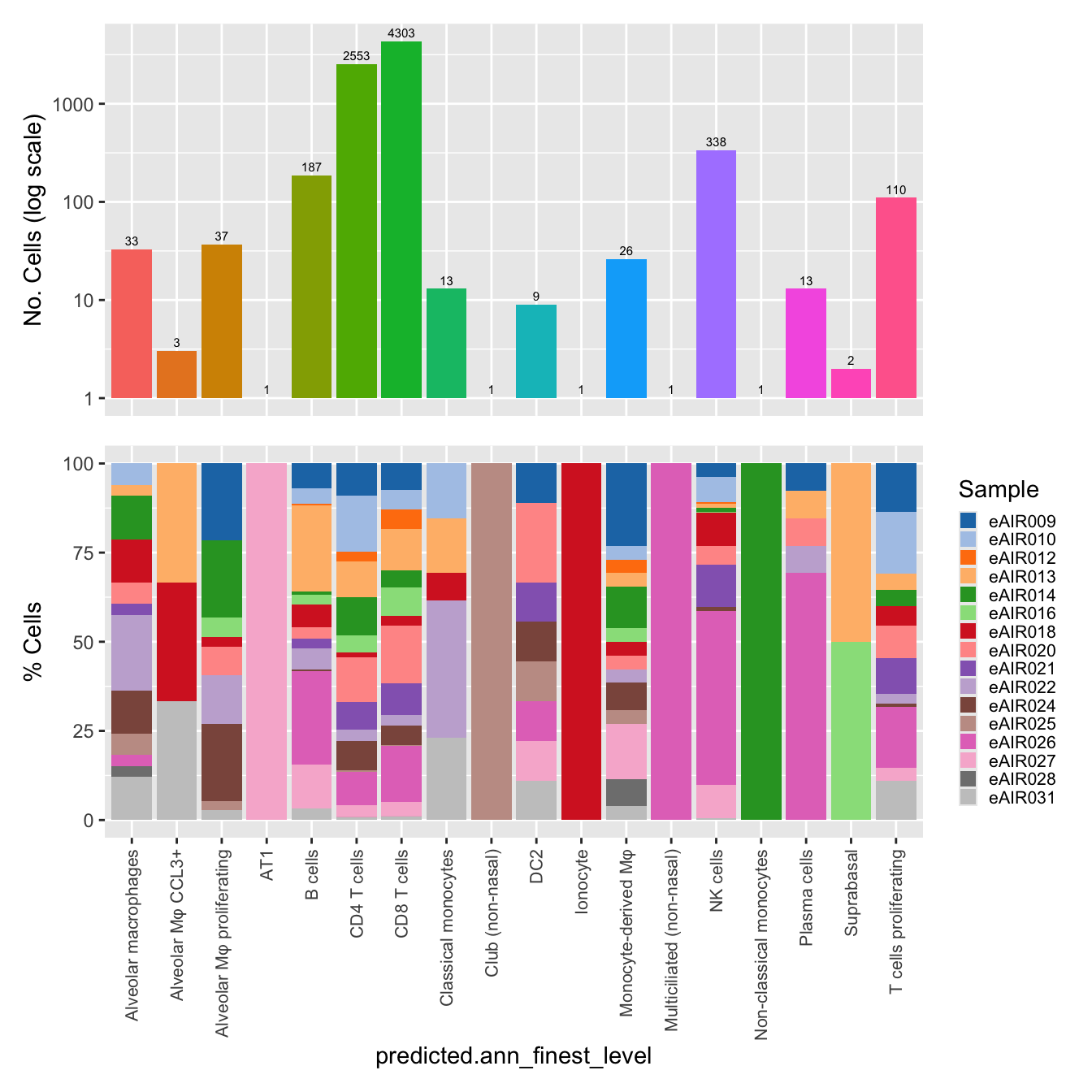
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
$cell_labels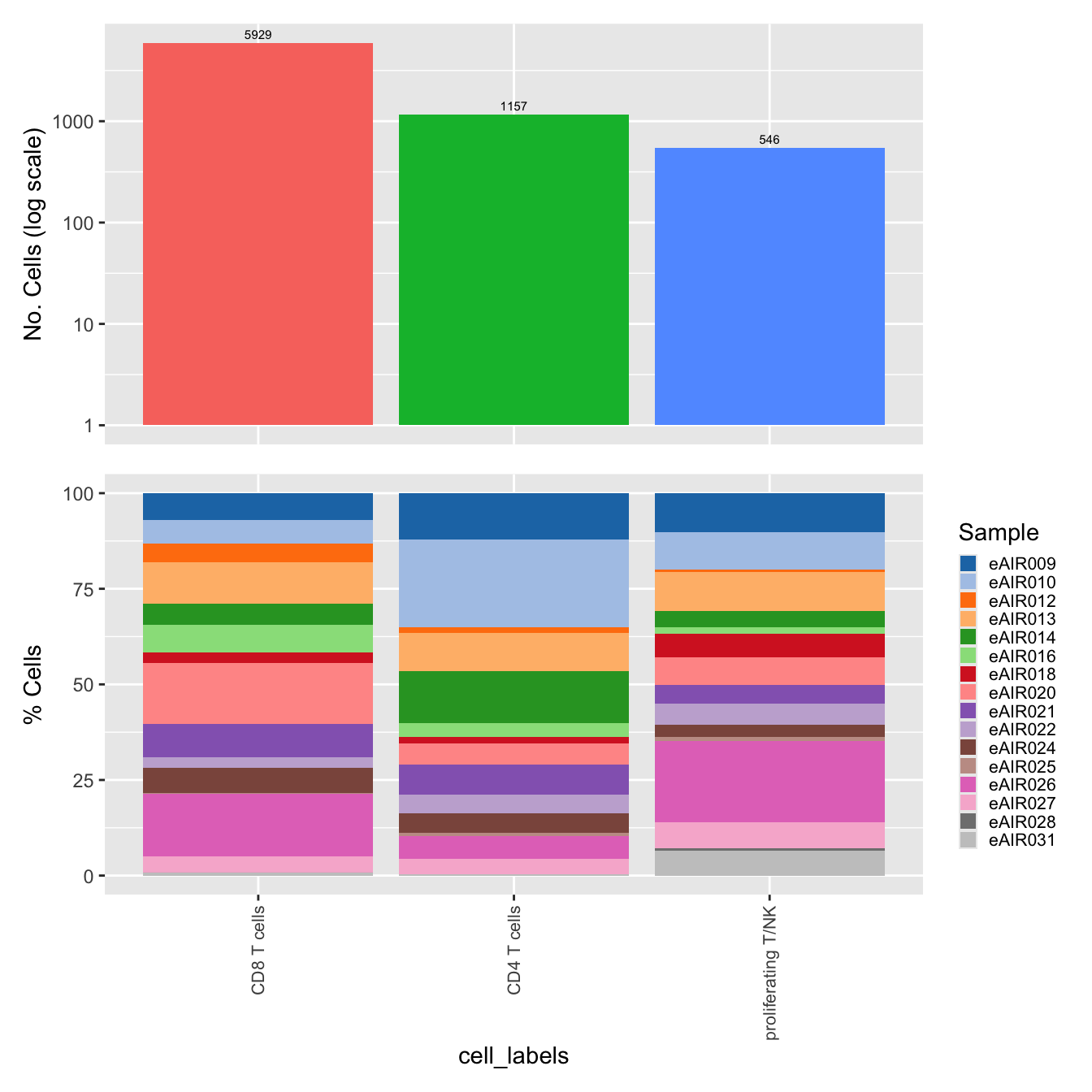
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
$cell_labels_v2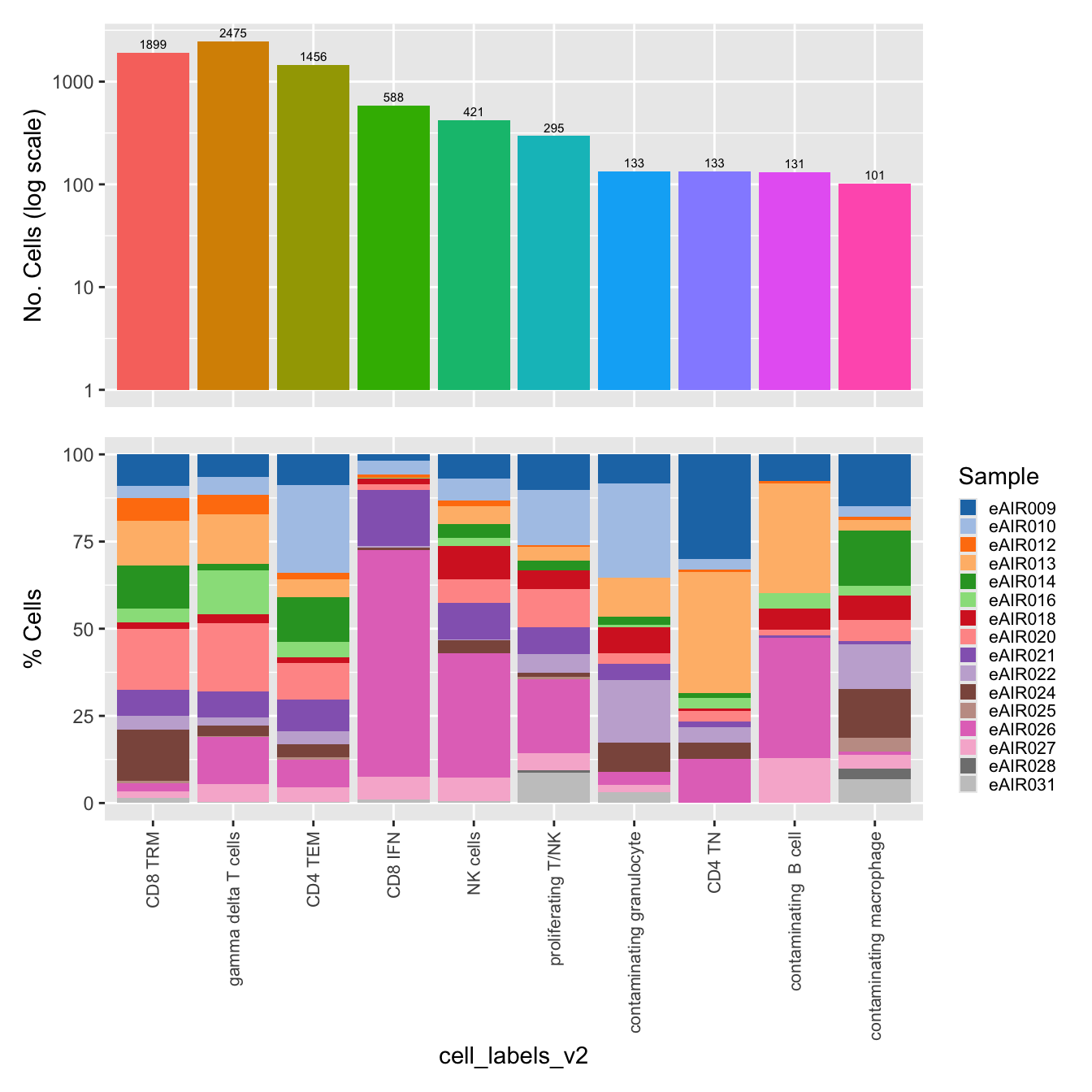
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
$RNA_snn_res.0.4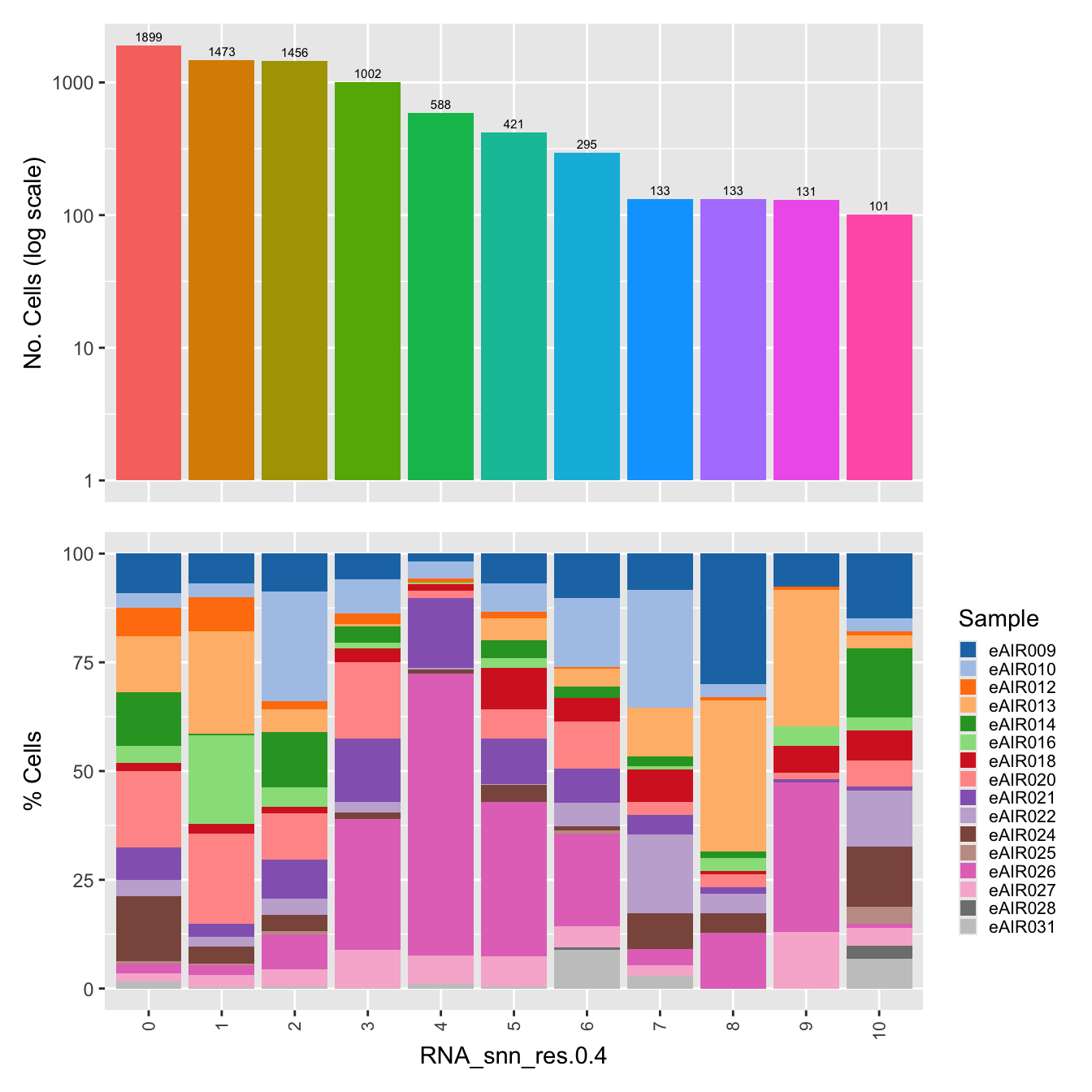
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
p1 <- paed_sub@meta.data %>%
dplyr::select(!!sym(opt_res), cell_labels_v2) %>% ggplot(aes(x = !!sym(opt_res),
fill = cell_labels_v2)) +
geom_bar() +
geom_text(aes(label = ..count..), stat = "count",
vjust = -0.5, colour = "black", size = 2) +
scale_y_log10() +
theme(axis.text.x = element_blank(),
axis.title.x = element_blank(),
axis.ticks.x = element_blank()) +
labs(y = "No. Cells (log scale)")
p2 <- paed_sub@meta.data %>%
dplyr::select(!!sym(opt_res), Sample) %>%
group_by(!!sym(opt_res), Sample) %>%
summarise(num = n()) %>%
mutate(prop = num / sum(num)) %>%
ggplot(aes(x = !!sym(opt_res), y = prop * 100,
fill = Sample)) +
geom_bar(stat = "identity") +
theme(axis.text.x = element_text(angle = 90,
vjust = 0.5,
hjust = 1,
size = 8)) +
labs(y = "% Cells", fill = "Sample") +
scale_fill_manual(values = combined_palette)`summarise()` has grouped output by 'RNA_snn_res.0.4'. You can override using
the `.groups` argument.# Combine the plots
(p1 / p2) & theme( legend.text = element_text(size = 8),
legend.key.size = unit(3, "mm"))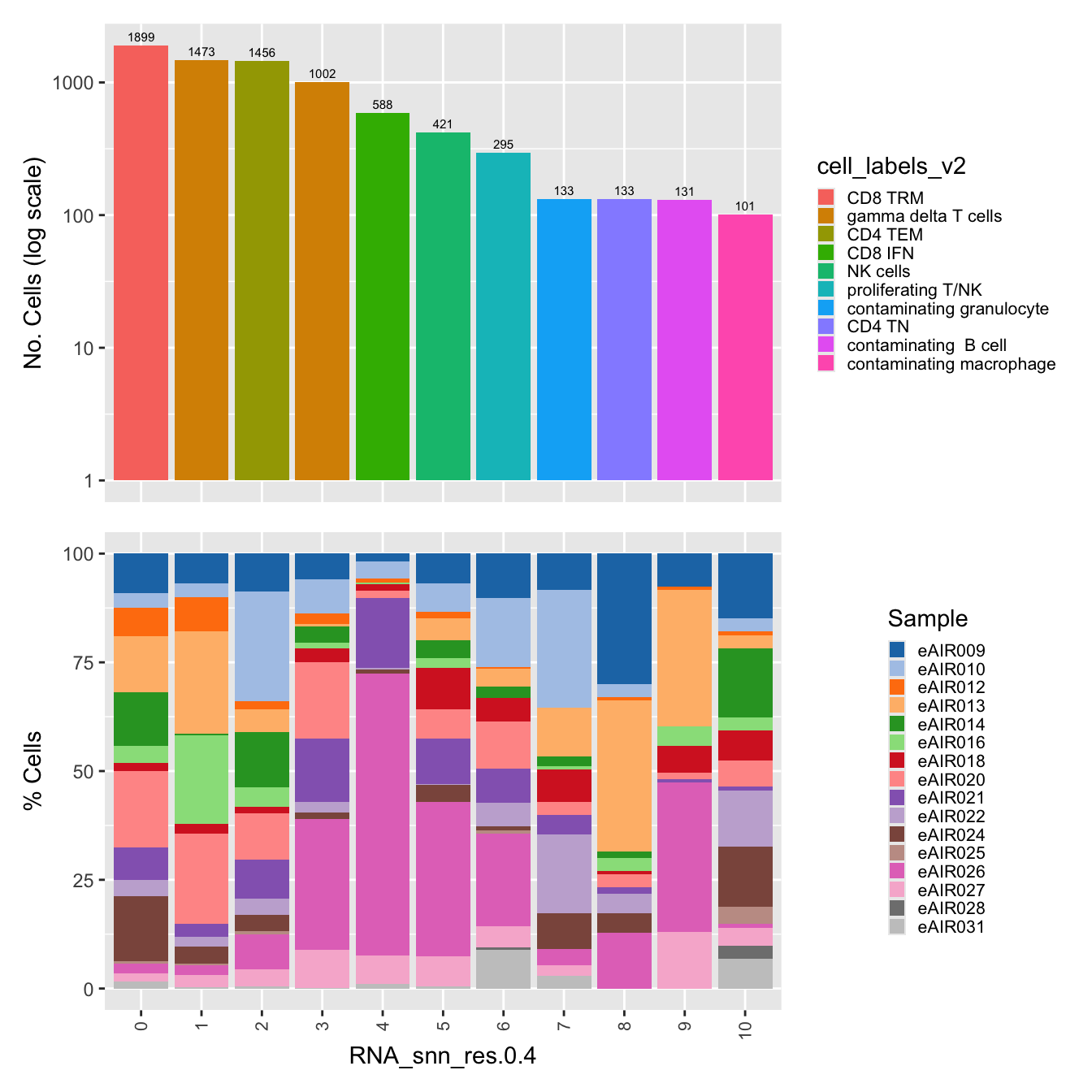
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
Additional QC plots for Proliferatink T/NK cluster
FeaturePlot(seu_obj, features=c("CD19", "MKI67"), reduction = 'umap', raster = FALSE, label = T, ncol = 2)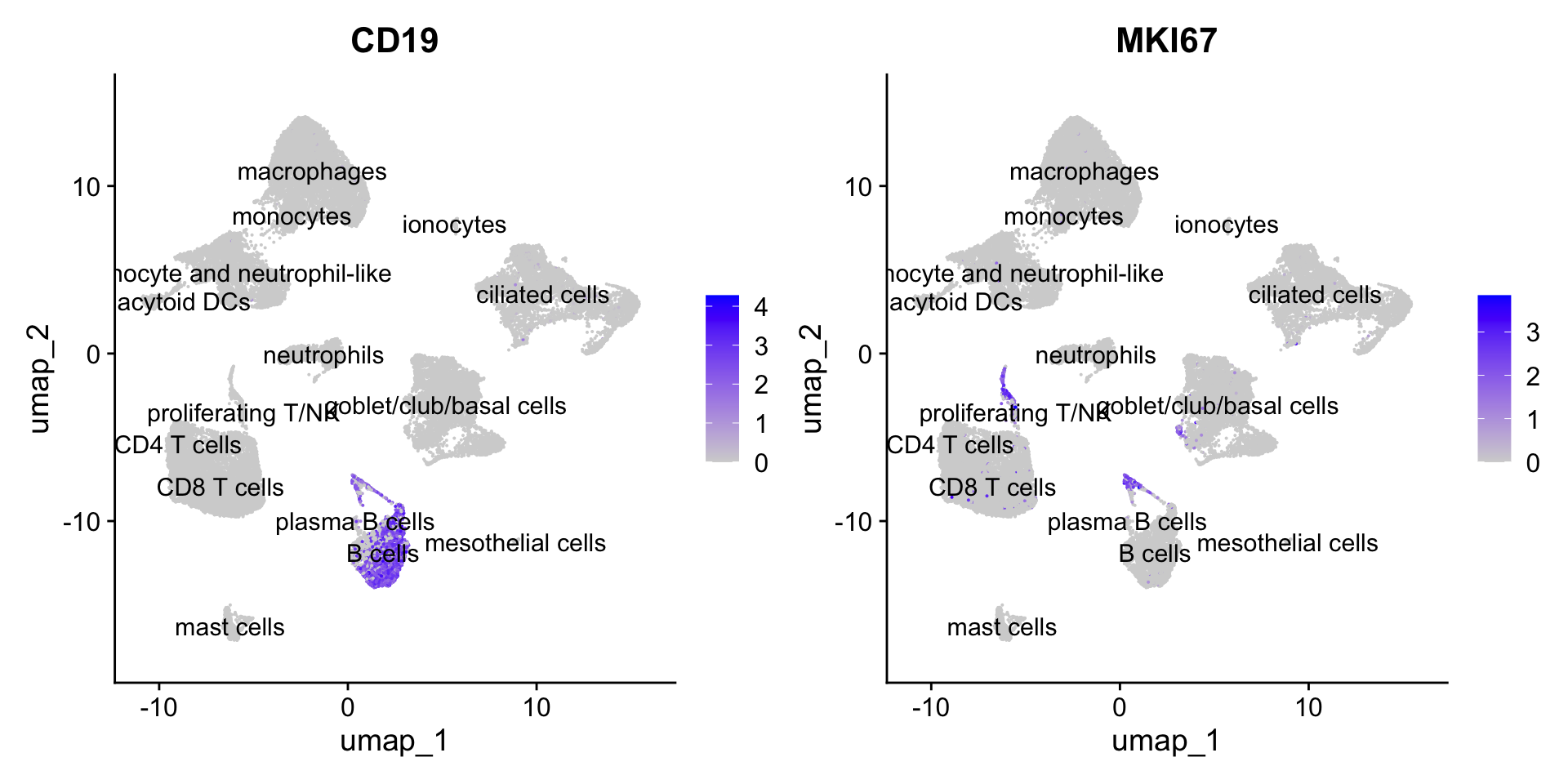
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
DimPlot(seu_obj, reduction = "umap", group.by = "predicted.ann_level_4", label = TRUE)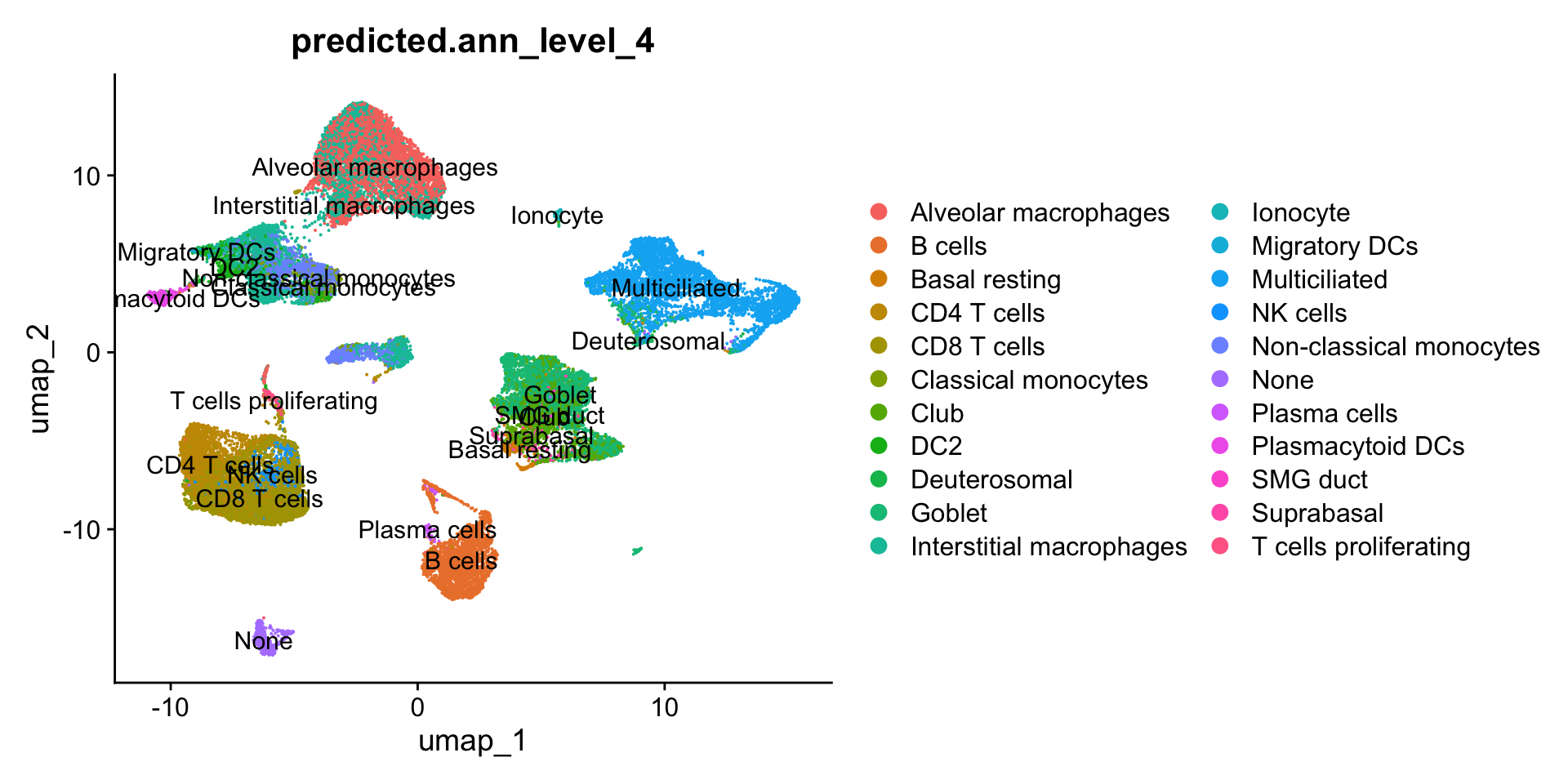
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
cl_13 <- as.data.frame(table(seu_obj$predicted.ann_level_4[seu_obj$RNA_snn_res.0.4 == "13"]))
colnames(cl_13) <- c("CellType", "Count")
cl_13 <- cl_13 %>% filter(Count > 0)
ggplot(cl_13, aes(x = reorder(CellType, Count), y = Count, fill = CellType)) +
geom_bar(stat = "identity") +
geom_text(aes(label = Count), hjust = -0.3) +
coord_flip() +
labs(title = "Azimuth (level4) cell types Cluster 13", x = "Cell Type", y = "Count") +
theme_minimal() +
theme(legend.position = "none") 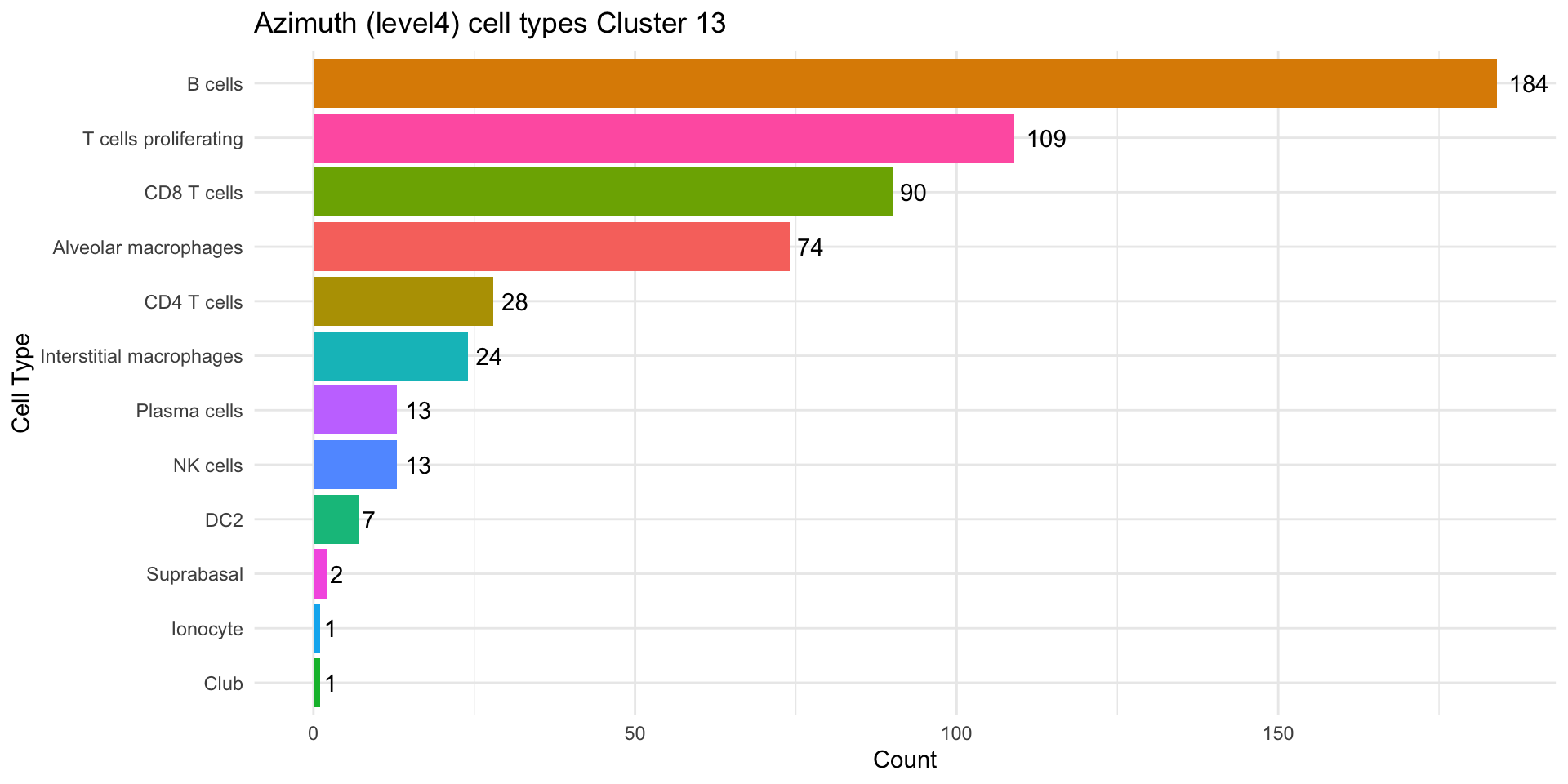
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
FeaturePlot(seu_obj, features = c('nCount_RNA',"nFeature_RNA"), pt.size=0.2, label = T, ncol =2)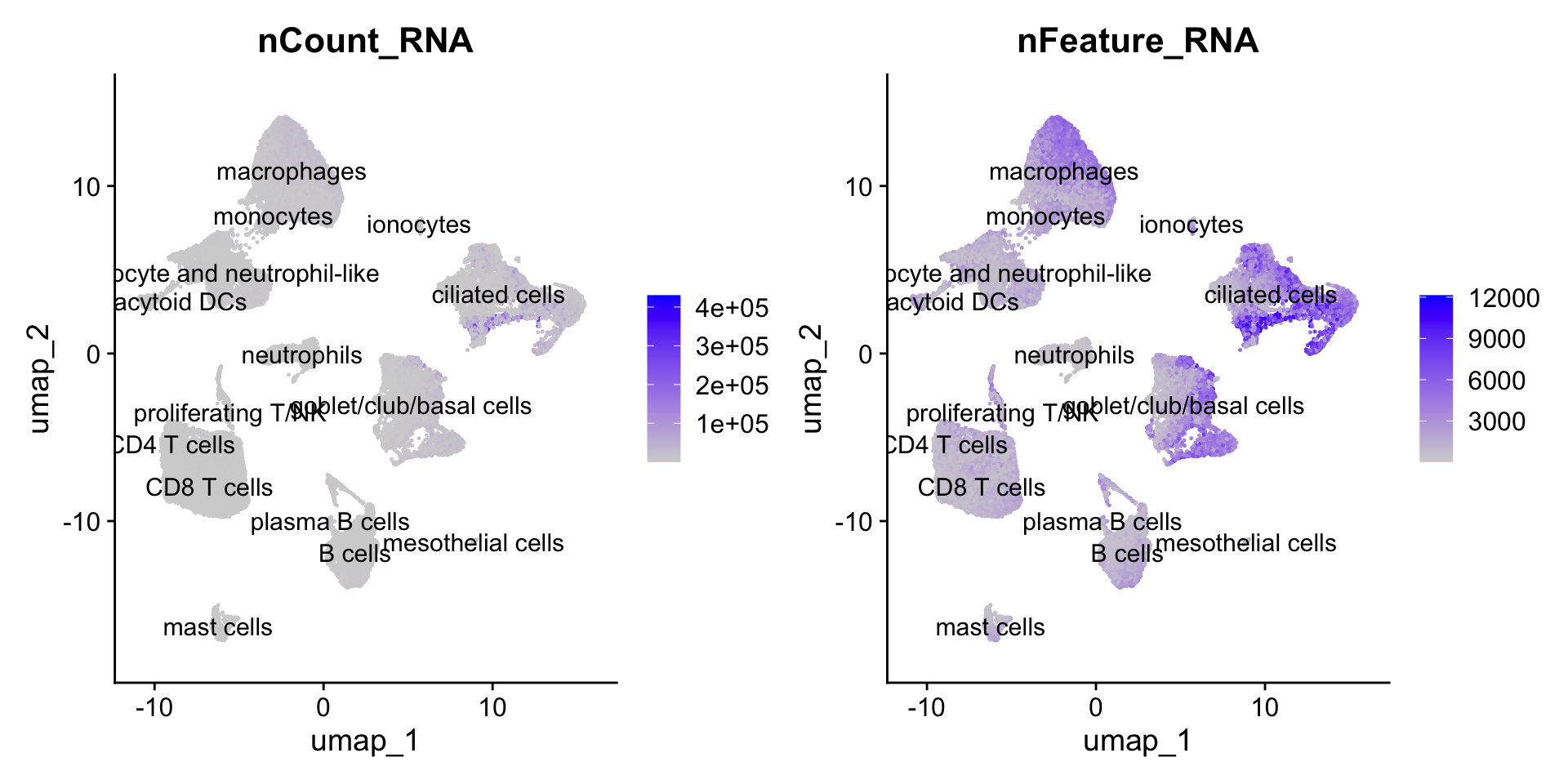
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
VlnPlot(seu_obj,features = c("nCount_RNA","nFeature_RNA"), pt.size = 0) &
theme(plot.title = element_text(size=10))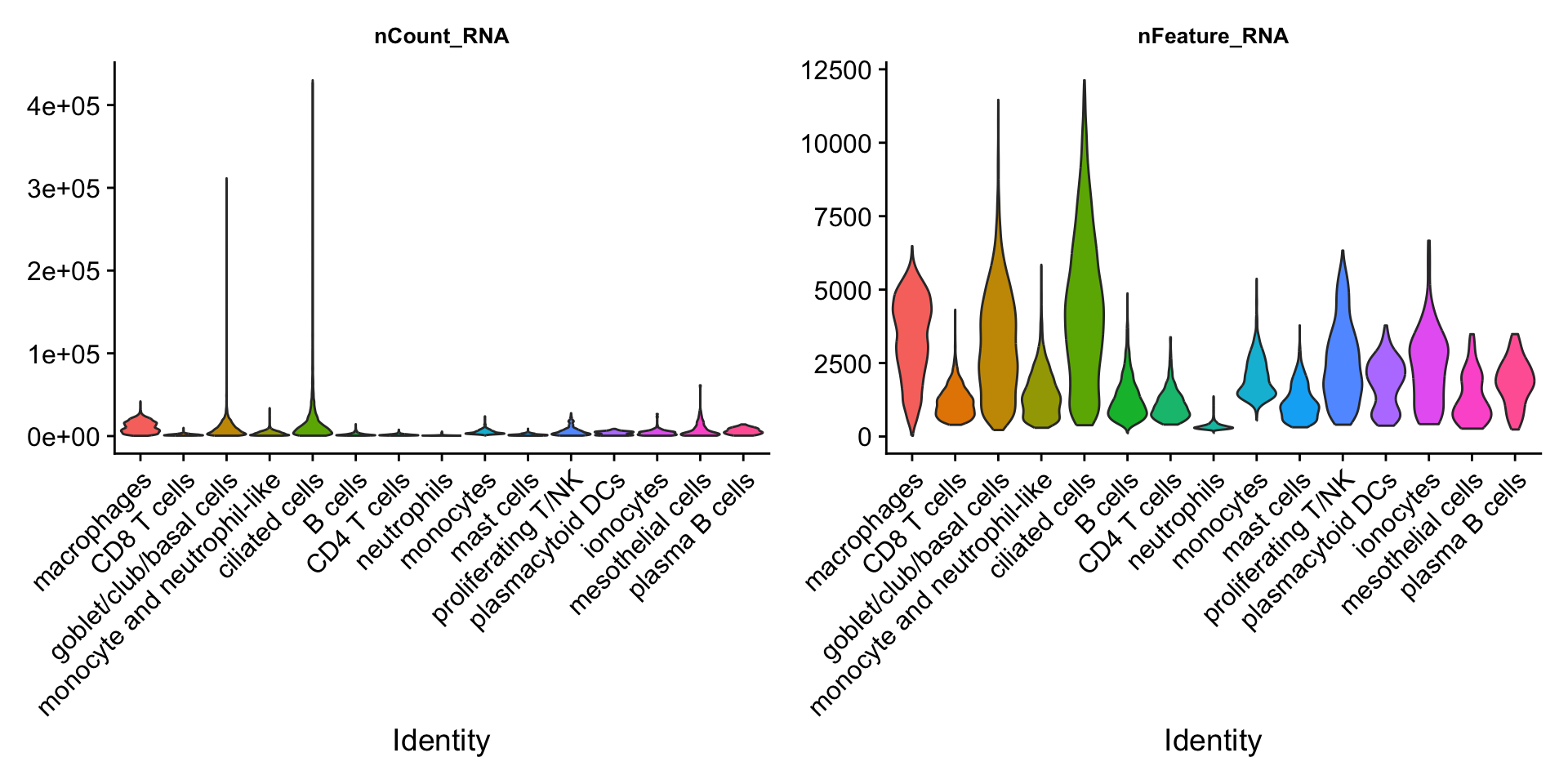
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
Save subclustered SEU object
out2 <- here("output",
"RDS", "AllBatches_Subclustering_SEUs", tissue,
paste0("G000231_Neeland_",tissue,".Tcell_population.subclusters.SEU.rds"))
#dir.create(out2)
if (!file.exists(out2)) {
saveRDS(paed_sub, file = out2)
}Other Clusters (excluding subclusters)
idx <- which(Idents(seu_obj) %in% c("CD4 T cells", "CD8 T cells", "proliferating T/NK"))
paed_sub <- seu_obj[,-idx]
paed_subAn object of class Seurat
18046 features across 26285 samples within 1 assay
Active assay: RNA (18046 features, 2000 variable features)
3 layers present: counts, data, scale.data
3 dimensional reductions calculated: pca, umap, umap.unintegratedlevels(paed_sub$cell_labels)[levels(paed_sub$cell_labels) == "goblet/club/basal cells"] <- "non-ciliated cells"
levels(Idents(paed_sub))[levels(Idents(paed_sub)) == "goblet/club/basal cells"] <- "non-ciliated cells"
paed_sub$cell_labels_v2 <- Idents(paed_sub)# Visualize the clustering results
DimPlot(paed_sub, reduction = "umap", group.by = "cluster", label = TRUE, label.size = 2.5, repel = TRUE, raster = FALSE )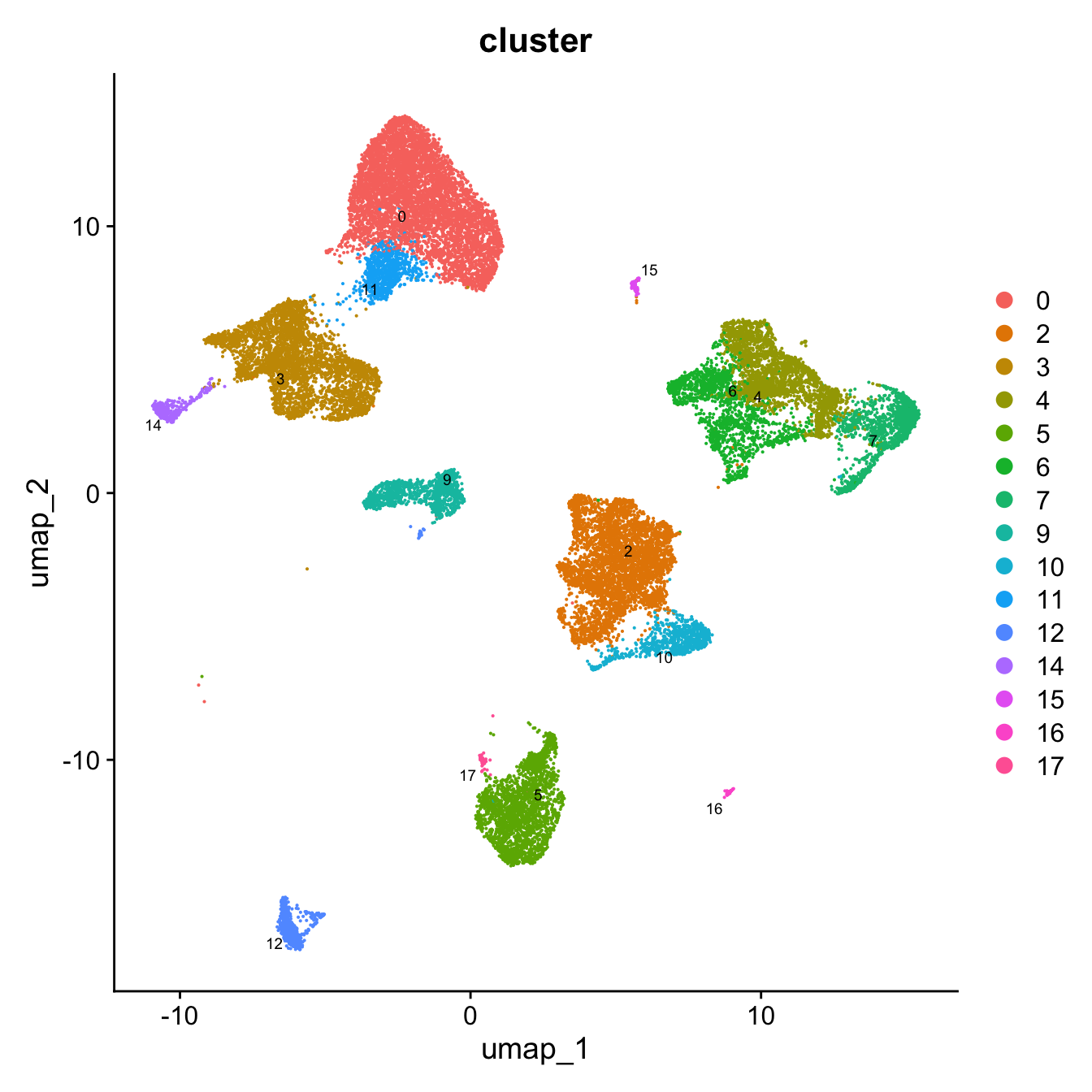
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
DimPlot(paed_sub, reduction = "umap", label = TRUE, label.size = 2.5, repel = TRUE, raster = FALSE )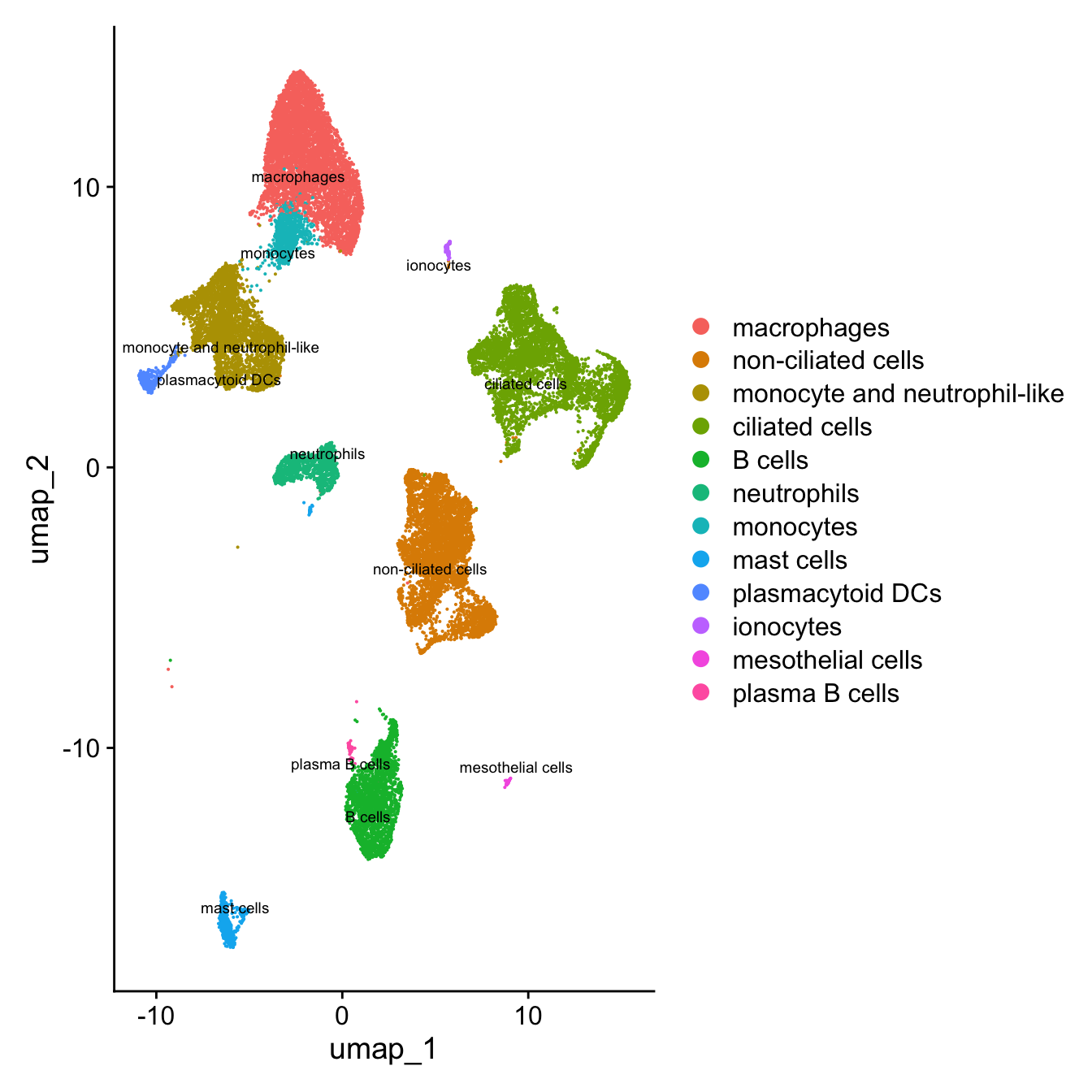
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
Save subclustered SEU object ( All other cells)
out2 <- here("output",
"RDS", "AllBatches_Subclustering_SEUs", tissue,
paste0("G000231_Neeland_",tissue,".all_other.subclusters.SEU.rds"))
#dir.create(out2)
if (!file.exists(out2)) {
saveRDS(paed_sub, file = out2)
}Merge seurat objects of subclusters
files <- list.files(here("output",
"RDS", "AllBatches_Subclustering_SEUs", tissue),
full.names = TRUE)
seuLst <- lapply(files, function(f) readRDS(f))
seu <- merge(seuLst[[1]],
y = seuLst[[2]])
seuAn object of class Seurat
18046 features across 33917 samples within 1 assay
Active assay: RNA (18046 features, 2000 variable features)
6 layers present: counts.1, counts.2, data.1, scale.data.1, data.2, scale.data.2Exploring the contaminated B cell cluster in T cells
idx <- which(Idents(seu) %in% c("B cells", "plasma B cells", "contaminating B cell"))
paed_bcells <- seu[,idx]
paed_bcells <- paed_bcells %>%
NormalizeData() %>%
FindVariableFeatures() %>%
ScaleData() %>%
RunPCA()Normalizing layer: counts.1Normalizing layer: counts.2Finding variable features for layer counts.1Finding variable features for layer counts.2Centering and scaling data matrixPC_ 1
Positive: TNFRSF13B, CD44, ADGRE5, DUSP4, ITGAX, MPEG1, EMP3, KLF2, NR4A1, ZEB2
LITAF, SEMA7A, PREX1, NR4A2, BHLHE40, VIM, TSC22D3, ZBTB32, JUNB, IER5
TYMP, FCRL4, RIN3, KDM6B, SOCS3, CCR6, CCDC50, FOSL2, NLRC5, FCRL5
Negative: MKI67, MEF2B, KIFC1, HIST1H1B, HMCES, AURKB, DTX1, BIRC5, TOP2A, MYBL1
UHRF1, NUGGC, TK1, STMN1, CDK1, MYBL2, TPX2, HMGB2, BUB1, HJURP
E2F2, MARCKSL1, TYMS, CDT1, DCAF12, AFF2, TCL1A, NUSAP1, HIST1H2BH, CCNB2
PC_ 2
Positive: IGHM, IGHD, NIBAN3, CD72, P2RX5, TCL1A, MMP17, PNPLA7, FCMR, OBSCN
FCER2, MYO15B, CXCR4, FOXP1, CR2, TREML2, KLHL14, TENT5C, VPREB3, MARCKSL1
NEIL1, CD200, CRIP3, KCNG1, DIPK1B, C16orf74, LY9, CNR2, TP53INP1, HAPLN3
Negative: FCRL4, ITGAX, CCR1, KCTD12, CCR5, GSN, ITGB7, TESC, BHLHE40, IL2RB
ADGRG5, RAB31, HCK, C12orf75, BHLHE41, VIM, PLD4, SOX5, IVNS1ABP, TPM4
MYO1F, PEA15, FCRL5, EFHD2, ZYX, PREX1, CMTM6, SEMA7A, SIGLEC6, RUNX2
PC_ 3
Positive: JCHAIN, CHPF, XBP1, MZB1, DERL3, AQP3, CKAP4, FNDC3B, ERN1, PRDM1
ITM2C, HID1, TXNDC5, SDF2L1, SSR4, HSP90B1, FKBP11, SEC11C, SELENOS, MANF
RRBP1, WFS1, CRELD2, HM13, MAN1A1, HPGD, NT5DC2, IGHA1, HYOU1, SELPLG
Negative: LTB, CD83, CXCR4, SPIB, NR4A1, CIITA, KDM6B, NFKBID, NR4A3, EGR3
KIFC1, NR4A2, METRNL, AURKB, ADGRE5, IER5, ITGAX, DUSP4, FOSL2, MKI67
TPX2, ARHGAP9, INF2, CDC20, BIRC5, KLF6, TOP2A, DUSP2, HIST1H1B, CDK1
PC_ 4
Positive: TLR10, TRIM22, PLAC8, SIGLEC14, XAF1, STAT1, IFI44L, FCRL1, CCR6, SAMD9L
CIITA, RESF1, GAPT, SELL, HHEX, CMPK2, CHI3L2, EIF2AK2, OAS2, CD24
IFIT3, IFI44, DOCK10, OAS1, MX1, TNFSF10, FCRL2, MX2, SLFN5, CD53
Negative: KDM6B, FOSL2, NR4A3, NFKBID, NR4A1, DUSP4, SRGN, SLC7A5, PIM3, NR4A2
EGR3, DUSP2, IRF4, LMNA, PER1, GRASP, FOSB, TRAF1, ADGRE5, SLCO4A1
NINJ1, CD83, CD70, JUNB, TXNDC5, MYC, CREM, RGCC, BCL2L11, BICDL1
PC_ 5
Positive: ISG15, IFI6, OAS3, IFIT1, IRF7, APOL6, CMPK2, LY6E, USP18, IFI44L
XAF1, MX1, WARS, IFIT3, MX2, IFIT2, TRIM22, OAS2, IFI44, RSAD2
GBP1, STAT1, HAPLN3, UBE2L6, STAT2, GBP4, HELZ2, HERC5, LAP3, IFI35
Negative: FCRL2, FCRL4, KCTD12, IGHA1, MGAT4A, METTL7A, CD180, MYL9, RFLNB, ADGRG5
CD27, CAMK1, IGHA2, MS4A7, DERL3, LGALS1, SMIM14, TNFRSF17, CLIP2, ITGB7
IFNGR1, RHOQ, RUNX2, MAML3, TLR10, CMTM3, LTB, APCDD1, HHEX, SNX22 paed_bcells <- RunUMAP(paed_bcells, dims = 1:30, reduction = "pca", reduction.name = "umap.bcell")10:50:45 UMAP embedding parameters a = 0.9922 b = 1.112Found more than one class "dist" in cache; using the first, from namespace 'spam'Also defined by 'BiocGenerics'10:50:45 Read 2671 rows and found 30 numeric columns10:50:45 Using Annoy for neighbor search, n_neighbors = 30Found more than one class "dist" in cache; using the first, from namespace 'spam'Also defined by 'BiocGenerics'10:50:45 Building Annoy index with metric = cosine, n_trees = 500% 10 20 30 40 50 60 70 80 90 100%[----|----|----|----|----|----|----|----|----|----|**************************************************|
10:50:45 Writing NN index file to temp file /var/folders/q8/kw1r78g12qn793xm7g0zvk94x2bh70/T//RtmpS8etyp/file624d707c805c
10:50:45 Searching Annoy index using 1 thread, search_k = 3000
10:50:46 Annoy recall = 100%
10:50:46 Commencing smooth kNN distance calibration using 1 thread with target n_neighbors = 30
10:50:47 Initializing from normalized Laplacian + noise (using RSpectra)
10:50:47 Commencing optimization for 500 epochs, with 111186 positive edges
10:50:49 Optimization finishedDimPlot(paed_bcells, group.by = "cell_labels_v2",raster = FALSE, repel = TRUE, label = TRUE, label.size = 4.5)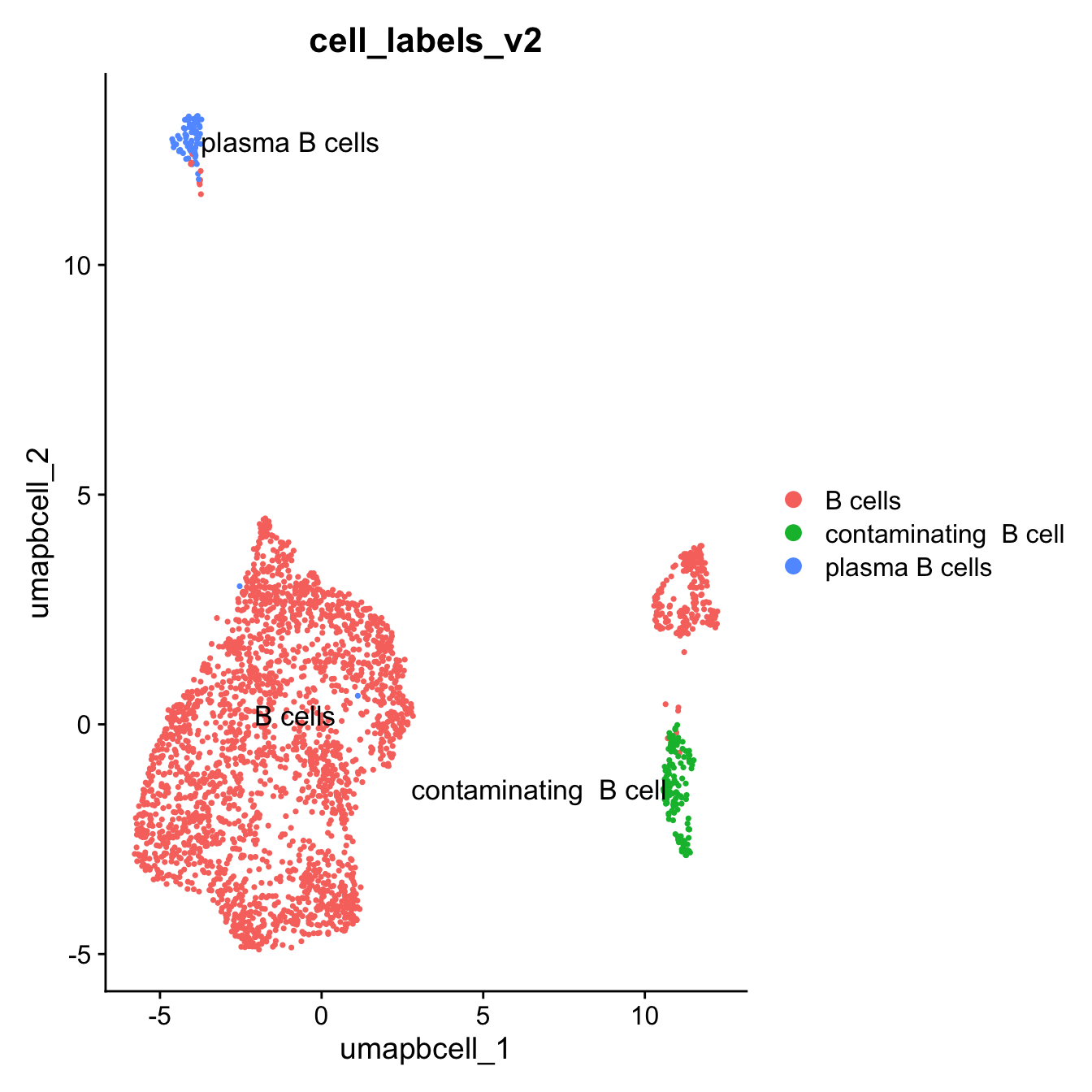
| Version | Author | Date |
|---|---|---|
| 07af966 | Gunjan Dixit | 2024-09-25 |
DimPlot(paed_bcells,raster = FALSE, repel = TRUE, label = TRUE, label.size = 4.5)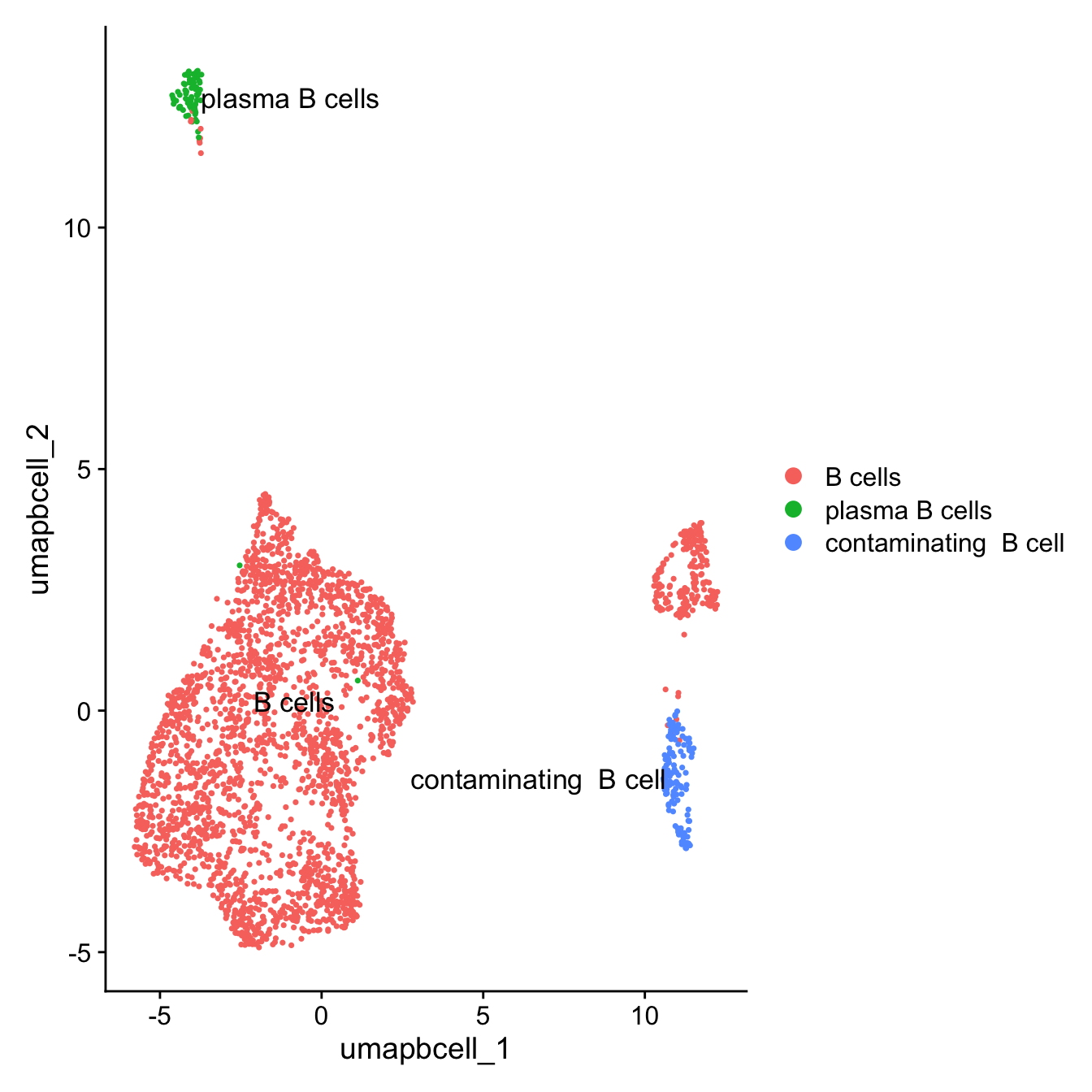
Reclustering B cells
The marker genes for this reclustering can be found here-
meta_data_columns <- colnames(paed_bcells@meta.data)
columns_to_remove <- grep("^RNA_snn_res", meta_data_columns, value = TRUE)
paed_bcells@meta.data <- paed_bcells@meta.data[, !(colnames(paed_bcells@meta.data) %in% columns_to_remove)]
resolutions <- seq(0.1, 1, by = 0.1)
paed_bcells <- FindNeighbors(paed_bcells, reduction = "pca", dims = 1:30)
paed_bcells <- FindClusters(paed_bcells, resolution = resolutions, algorithm = 3)Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 2671
Number of edges: 100594
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.9315
Number of communities: 4
Elapsed time: 1 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 2671
Number of edges: 100594
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.8899
Number of communities: 5
Elapsed time: 0 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 2671
Number of edges: 100594
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.8659
Number of communities: 7
Elapsed time: 0 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 2671
Number of edges: 100594
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.8494
Number of communities: 8
Elapsed time: 0 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 2671
Number of edges: 100594
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.8349
Number of communities: 8
Elapsed time: 0 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 2671
Number of edges: 100594
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.8210
Number of communities: 9
Elapsed time: 0 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 2671
Number of edges: 100594
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.8086
Number of communities: 10
Elapsed time: 0 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 2671
Number of edges: 100594
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.7982
Number of communities: 11
Elapsed time: 0 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 2671
Number of edges: 100594
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.7878
Number of communities: 12
Elapsed time: 0 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 2671
Number of edges: 100594
Running smart local moving algorithm...
Maximum modularity in 10 random starts: 0.7775
Number of communities: 12
Elapsed time: 0 secondsDimHeatmap(paed_bcells, dims = 1:10, cells = 500, balanced = TRUE)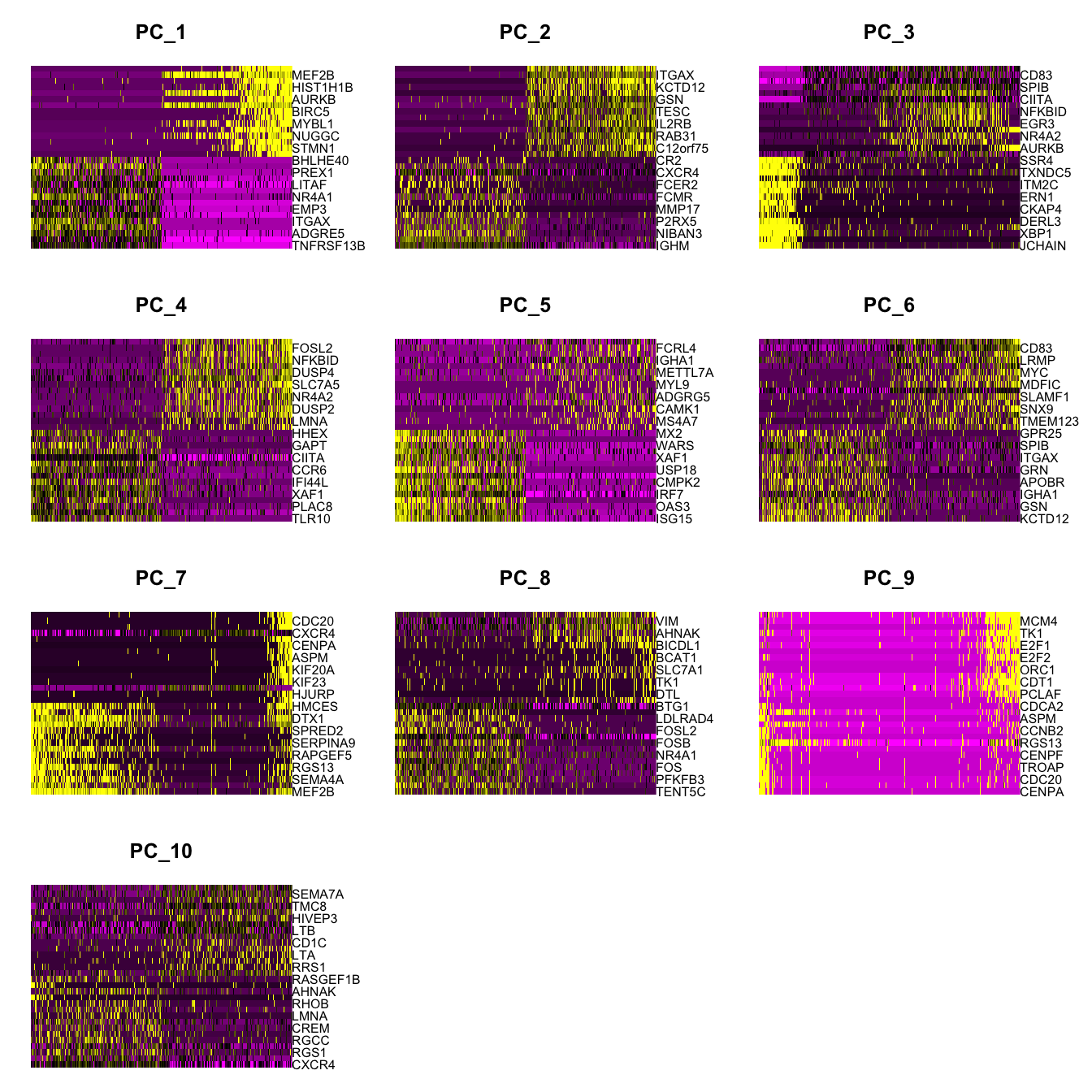
clustree(paed_bcells, prefix = "RNA_snn_res.")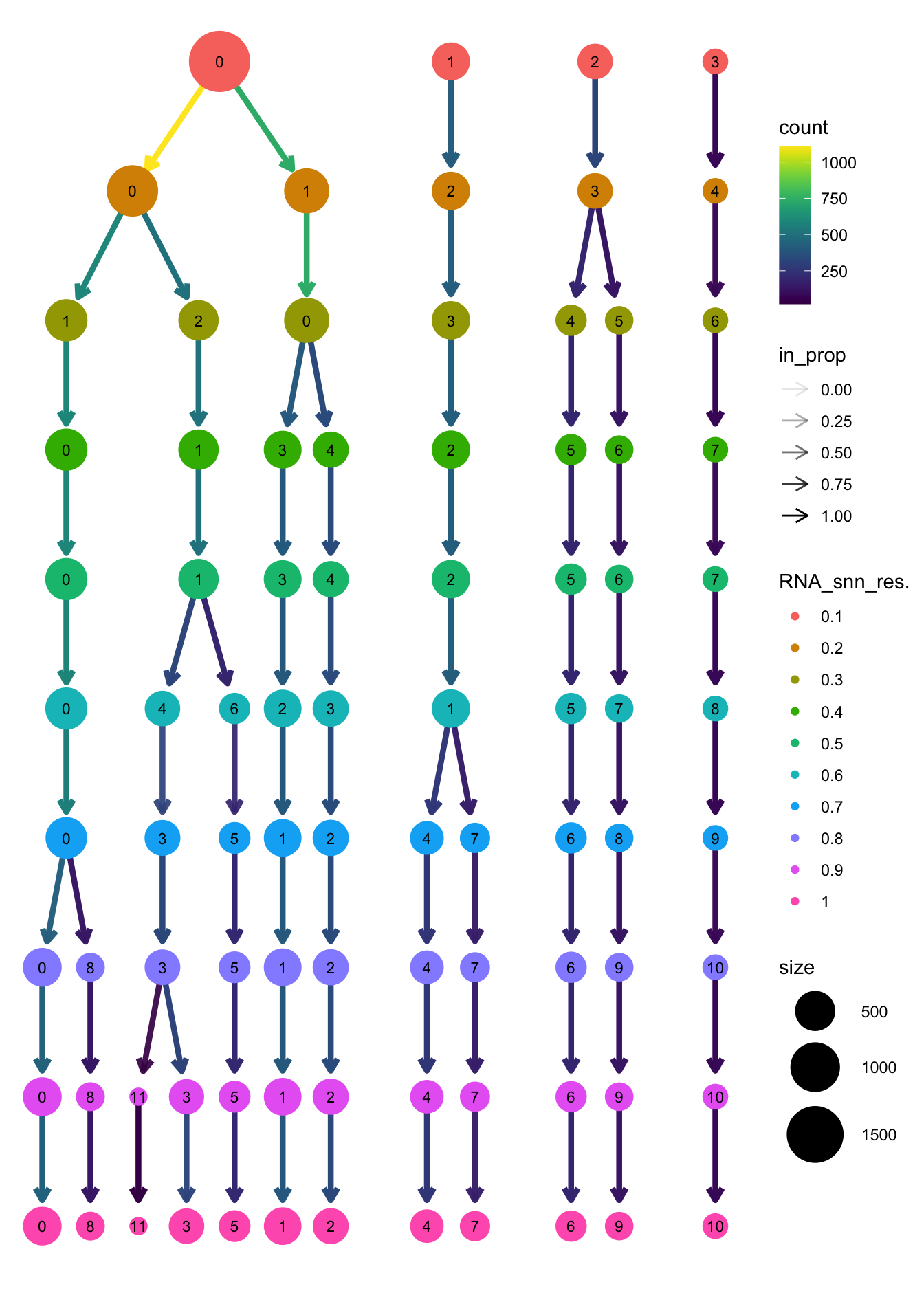
opt_res <- "RNA_snn_res.0.3"
n <- nlevels(paed_bcells$RNA_snn_res.0.3)
paed_bcells$RNA_snn_res.0.3 <- factor(paed_bcells$RNA_snn_res.0.3, levels = seq(0,n-1))
paed_bcells$seurat_clusters <- NULL
Idents(paed_bcells) <- paed_bcells$RNA_snn_res.0.3DimPlot(paed_bcells, group.by = "RNA_snn_res.0.3", label = TRUE, label.size = 4.5, repel = TRUE, raster = FALSE )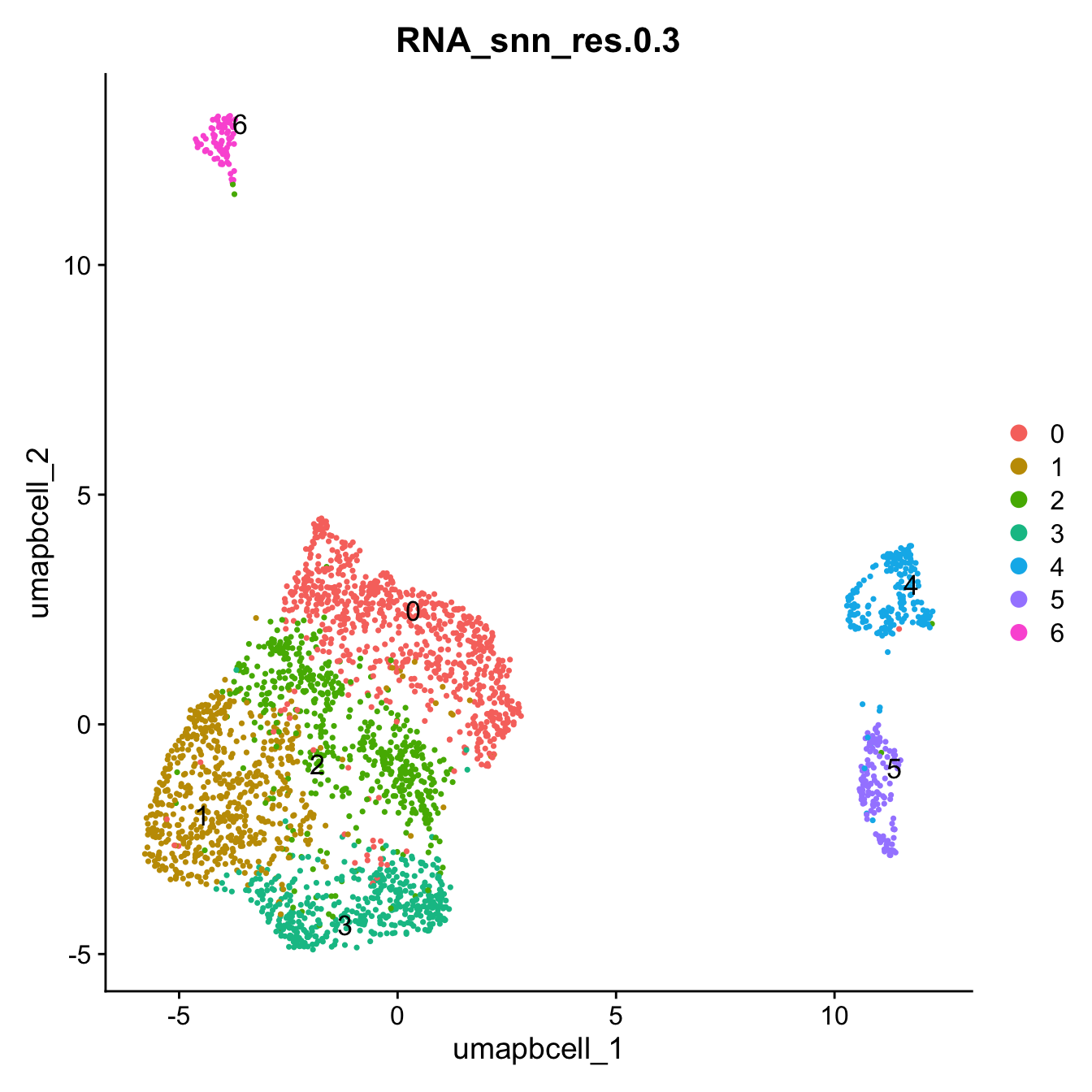
paed_bcells <- JoinLayers(paed_bcells)
paed_bcells.markers <- FindAllMarkers(paed_bcells, only.pos = TRUE, min.pct = 0.25, logfc.threshold = 0.25)Calculating cluster 0Calculating cluster 1Calculating cluster 2Calculating cluster 3Calculating cluster 4Calculating cluster 5Calculating cluster 6paed_bcells.markers %>%
group_by(cluster) %>% unique() %>%
top_n(n = 5, wt = avg_log2FC) -> top5
paed_bcells.markers %>%
group_by(cluster) %>%
slice_head(n=1) %>%
pull(gene) -> best.wilcox.gene.per.cluster
best.wilcox.gene.per.cluster[1] "IGHD" "KDM6B" "MS4A1" "FCRL4" "MEF2B" "UHRF1" "CHPF" FeaturePlot(paed_bcells,features=best.wilcox.gene.per.cluster, raster = FALSE, label = T, ncol = 3)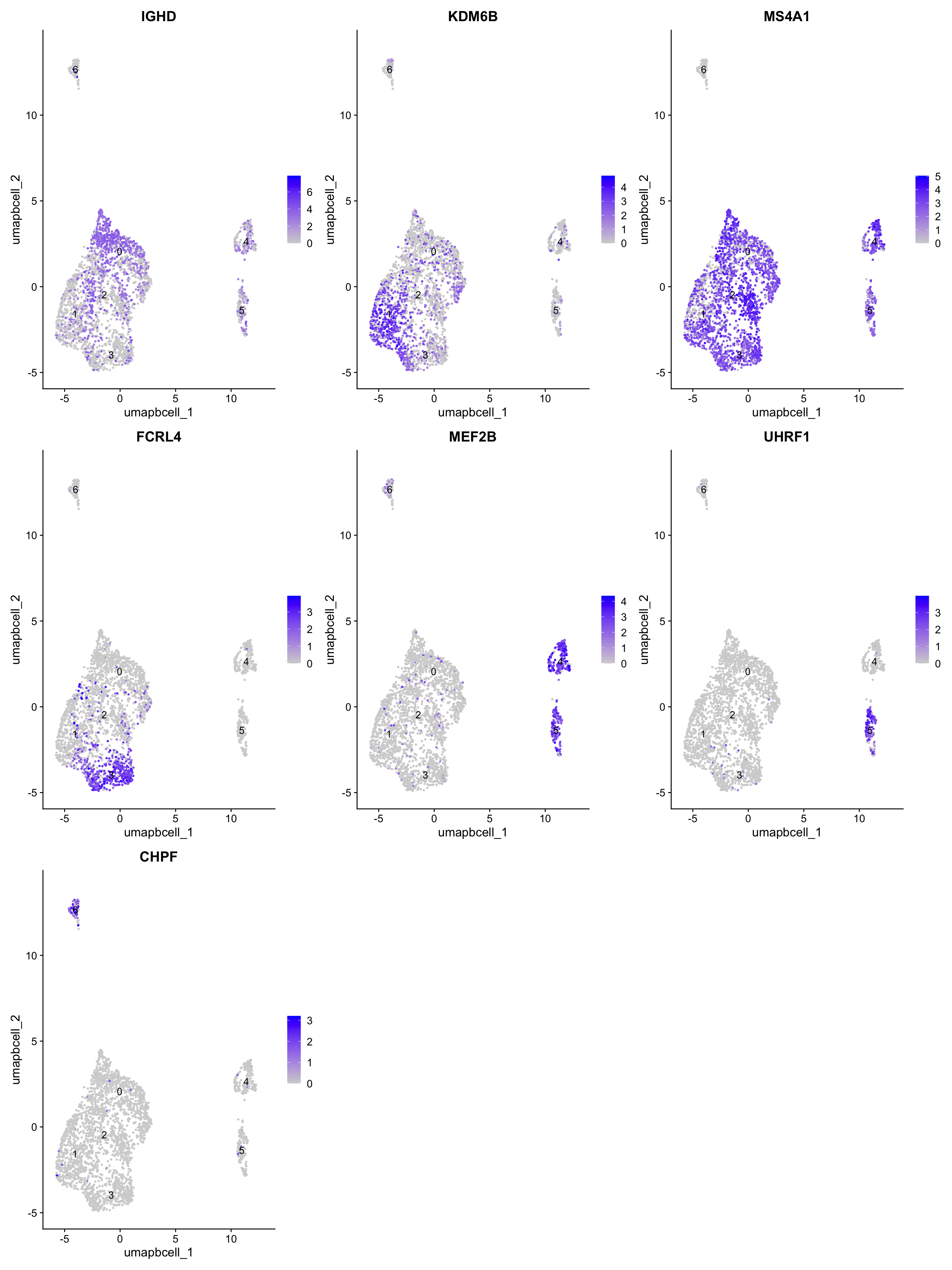
Top 10 marker genes from Seurat
## Seurat top markers
top10 <- paed_bcells.markers %>%
group_by(cluster) %>%
top_n(n = 10, wt = avg_log2FC) %>%
ungroup() %>%
distinct(gene, .keep_all = TRUE) %>%
arrange(cluster, desc(avg_log2FC))
cluster_colors <- paletteer::paletteer_d("pals::glasbey")[factor(top10$cluster)]
DotPlot(paed_bcells,
features = unique(top10$gene),
group.by = opt_res,
cols = c("azure1", "blueviolet"),
dot.scale = 3, assay = "RNA") +
RotatedAxis() +
FontSize(y.text = 8, x.text = 12) +
labs(y = element_blank(), x = element_blank()) +
coord_flip() +
theme(axis.text.y = element_text(color = cluster_colors)) +
ggtitle("Top 10 marker genes per cluster (Seurat)")Warning: Vectorized input to `element_text()` is not officially supported.
ℹ Results may be unexpected or may change in future versions of ggplot2.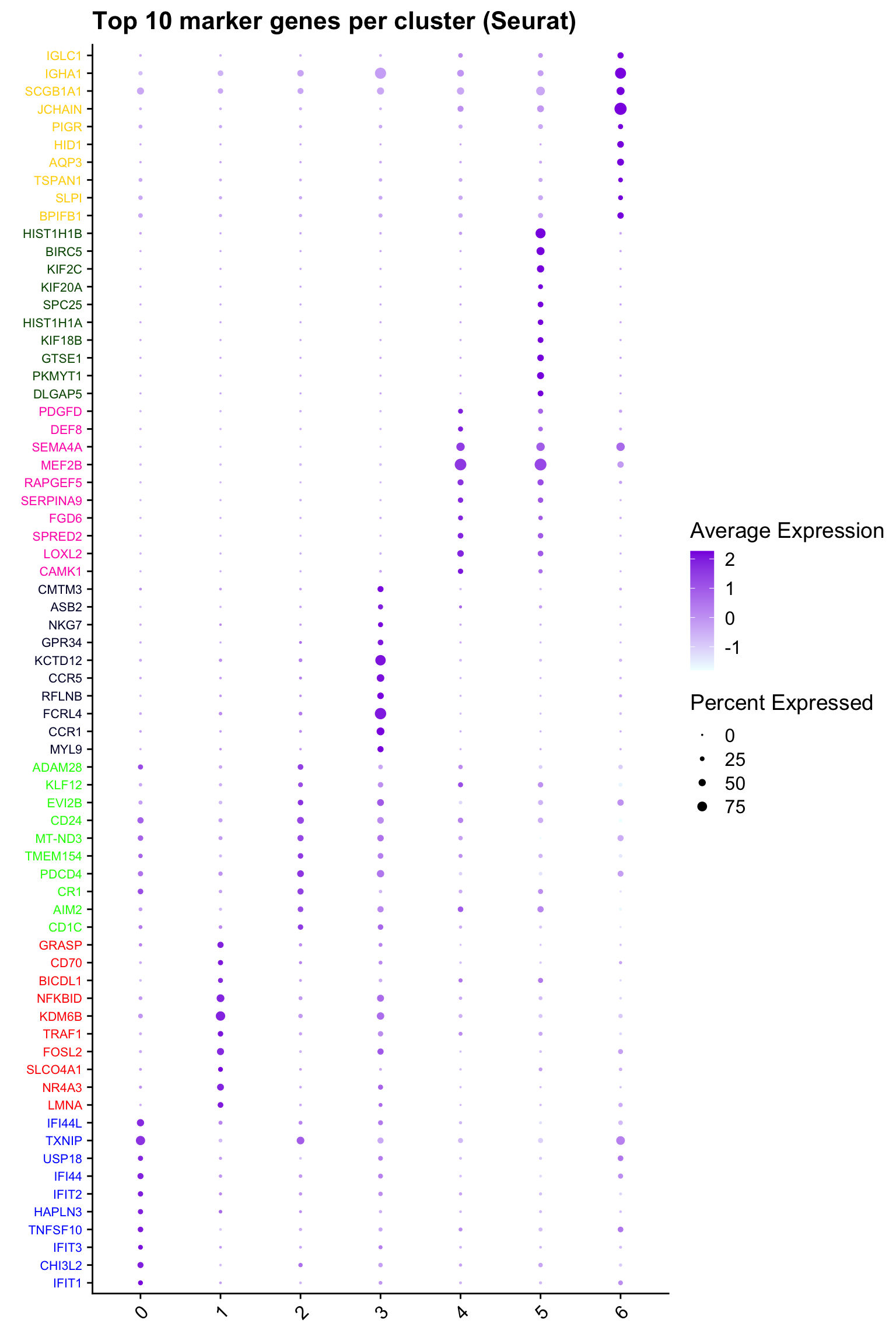
out_markers <- here("output",
"CSV",
paste(tissue,"_Marker_genes_Reclustered_Bcell_population.",opt_res, sep = ""))
dir.create(out_markers, recursive = TRUE, showWarnings = FALSE)
for (cl in unique(paed_bcells.markers$cluster)) {
cluster_data <- paed_bcells.markers %>% dplyr::filter(cluster == cl)
file_name <- here(out_markers, paste0("G000231_Neeland_",tissue, "_cluster_", cl, ".csv"))
if (!file.exists(file_name)) {
write.csv(cluster_data, file = file_name)
}
}Excluding contaminating labels
idx <- which(grepl("^contaminating", Idents(seu)))
seu <- seu[, -idx]merged <- seu %>%
NormalizeData() %>%
FindVariableFeatures() %>%
ScaleData() %>%
RunPCA()Normalizing layer: counts.1Normalizing layer: counts.2Finding variable features for layer counts.1Finding variable features for layer counts.2Centering and scaling data matrixPC_ 1
Positive: C9orf24, SNTN, C20orf85, PIFO, RSPH1, ZMYND10, FOXJ1, LRRC46, SPATA18, TEKT1
DNALI1, FAM92B, ROPN1L, LRRC10B, C5orf49, RSPH4A, AC007906.2, CAPSL, C11orf88, FAM216B
C9orf116, DRC3, B9D1, LDLRAD1, EFCAB1, ERICH3, WDR66, HYDIN, SAXO2, VWA3A
Negative: VIM, ZEB2, SPI1, PFN1, THEMIS2, COTL1, FCER1G, TYROBP, IFI30, FGR
IRF8, CD68, CD83, LGALS1, ALOX5, GRN, ITGAX, EMILIN2, METRNL, HCK
CYBB, MS4A7, AIF1, CXCR4, FCGR3A, TFRC, LYZ, PHACTR1, C1QB, HAVCR2
PC_ 2
Positive: KRT7, F3, CREB3L1, FAM3D, S100A16, PRSS8, MDK, CEACAM6, C3, SDC1
RDH10, MUC1, SLC6A14, BPIFB1, S100P, ALPL, WFDC2, SLC5A8, A4GALT, MUC5B
ASS1, VSIG2, LCN2, SERPINB3, TMPRSS4, UPK1B, GPC1, GPRC5A, FCGBP, CXCL6
Negative: VIM, KCTD12, SPI1, FCER1G, TYROBP, CD68, CTSZ, LGALS1, CD4, ZEB2
FGR, EMILIN2, MS4A7, THEMIS2, CD83, AIF1, IRF8, COTL1, CYBB, HCK
C1QB, S100A10, ALOX5, FCGR3A, IFI30, SLC15A3, TFRC, DSE, ITGAX, HMOX1
PC_ 3
Positive: CST3, LYZ, ALDH2, GRN, CEBPB, CD68, IFI30, SLC15A3, TYROBP, FCER1G
EMILIN2, MS4A7, LMNA, C1QB, CTSZ, TGM2, AIF1, HMOX1, S100A9, TYMP
RBM47, SPI1, FCGR3A, MS4A4A, RHOB, LGALS1, DSE, TFRC, HCK, AXL
Negative: IL32, CCL5, NKG7, PRF1, TRBC1, GZMA, LTB, CD69, HOPX, GNLY
TIGIT, CCND2, CD27, CST7, FASLG, CXCR4, CD5, ARL4C, ITM2C, IGHM
GZMB, PIM2, LAG3, P2RX5, CD79A, TCF7, HIST1H1C, LY9, KLRC1, TBX21
PC_ 4
Positive: CCL5, IL32, PRF1, GZMA, TRBC1, NKG7, GNLY, FASLG, CST7, TIGIT
ANXA1, KLRC1, TNFAIP3, CSF1, LAG3, CCL4, CD5, IL7R, HOPX, GZMH
MAF, TNFSF14, CRTAM, KLRB1, TCF7, NCR1, JUN, PHLDA1, TRDC, IFNG
Negative: CD79A, MS4A1, TNFRSF13C, CD19, PAX5, BANK1, SPIB, TNFRSF13B, CD79B, CD22
FCRL5, IGKC, WDFY4, POU2AF1, FCRLA, CXCR5, BLNK, TLR10, IGHG1, CLEC17A
PLD4, BLK, BCL11A, FCRL2, IGHM, POU2F2, TSPAN33, CIITA, MPEG1, IGHA1
PC_ 5
Positive: BHLHE41, C1QB, PPARG, RBP4, ALDH2, C8B, FABP5, FN1, TFRC, CRIP1
AGRP, LDHB, HSPB1, BCAR1, CA2, LMNA, CCL18, INF2, YBX1, STMN1
LY86, CHDH, TUBB, MS4A7, NME2, CAMP, PTMS, GAS2L3, MS4A4A, AXL
Negative: APOBEC3A, IDO1, ISG15, ISG20, RSAD2, MX2, SERPINB9, CXCL11, IRF7, LILRA5
CD300E, CXCL10, IFIT2, SOCS3, IFITM3, CSF3R, SPHK1, GBP1, SAT1, GBP5
IFIT1, MX1, IFI44L, CMPK2, NLRP3, CCL8, HELZ2, CALHM6, PFKFB3, OAS3 merged <- RunUMAP(merged, dims = 1:30, reduction = "pca", reduction.name = "umap.merged")10:51:25 UMAP embedding parameters a = 0.9922 b = 1.112Found more than one class "dist" in cache; using the first, from namespace 'spam'Also defined by 'BiocGenerics'10:51:25 Read 33552 rows and found 30 numeric columns10:51:25 Using Annoy for neighbor search, n_neighbors = 30Found more than one class "dist" in cache; using the first, from namespace 'spam'Also defined by 'BiocGenerics'10:51:25 Building Annoy index with metric = cosine, n_trees = 500% 10 20 30 40 50 60 70 80 90 100%[----|----|----|----|----|----|----|----|----|----|**************************************************|
10:51:27 Writing NN index file to temp file /var/folders/q8/kw1r78g12qn793xm7g0zvk94x2bh70/T//RtmpS8etyp/file624d52e5b43
10:51:27 Searching Annoy index using 1 thread, search_k = 3000
10:51:32 Annoy recall = 100%
10:51:33 Commencing smooth kNN distance calibration using 1 thread with target n_neighbors = 30
10:51:33 Initializing from normalized Laplacian + noise (using RSpectra)
10:51:39 Commencing optimization for 200 epochs, with 1419026 positive edges
10:51:49 Optimization finishedp4 <- DimPlot(merged, reduction = "umap.merged", group.by = "cell_labels_v2",raster = FALSE, repel = TRUE, label = TRUE, label.size = 4.5) + ggtitle(paste0(tissue, ": UMAP with annotations")) + NoLegend()
p4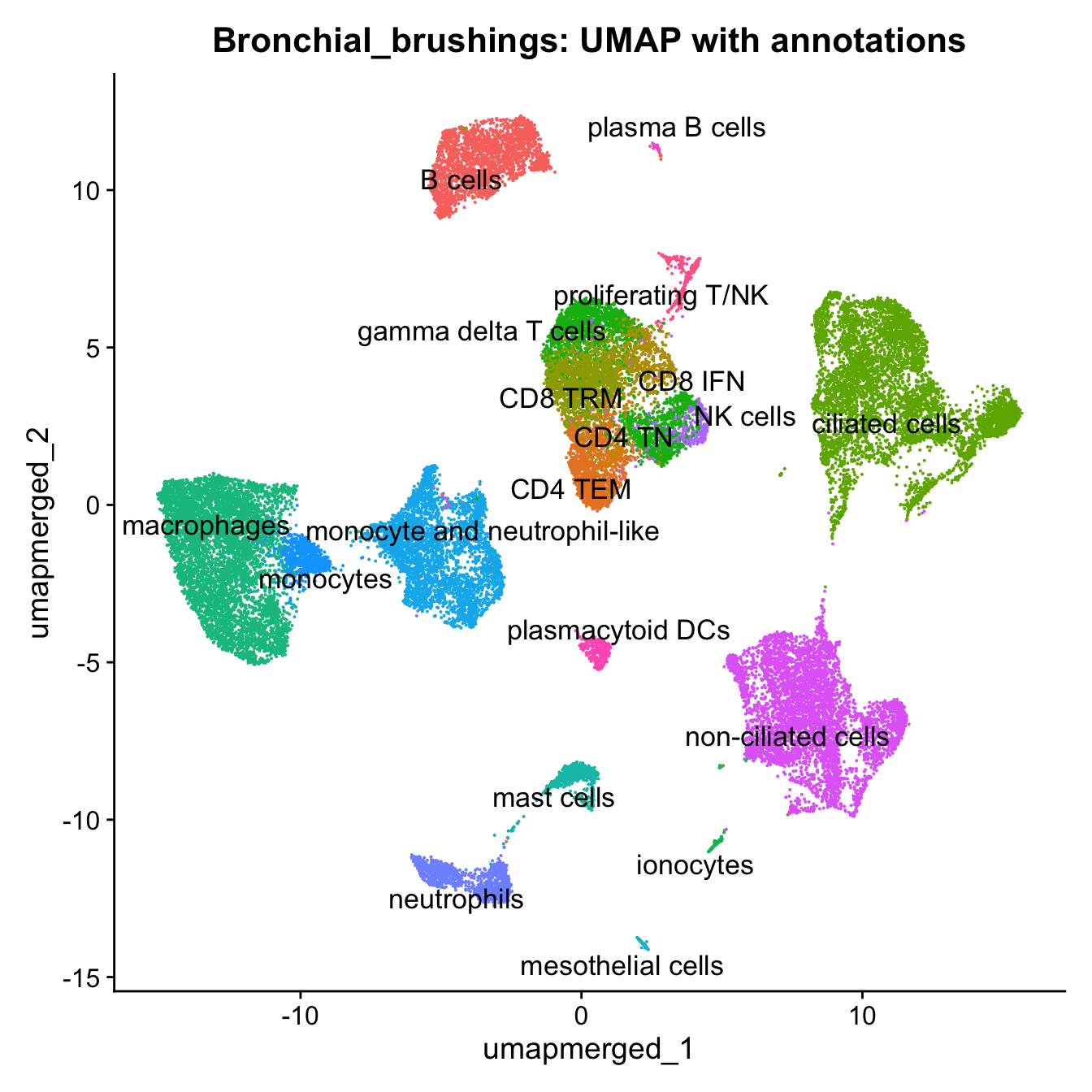
Save Final SEU object ( All cells)
out3 <- here("output",
"RDS", "AllBatches_Final_Clusters_SEUs",
paste0("G000231_Neeland_",tissue,".final_clusters.SEU.rds"))
if (!file.exists(out3)) {
saveRDS(merged, file = out3)
}Session Info
sessioninfo::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.3.2 (2023-10-31)
os macOS Sonoma 14.6.1
system aarch64, darwin20
ui X11
language (EN)
collate en_US.UTF-8
ctype en_US.UTF-8
tz Australia/Melbourne
date 2024-09-26
pandoc 3.1.1 @ /Users/dixitgunjan/Desktop/RStudio.app/Contents/Resources/app/quarto/bin/tools/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
abind 1.4-5 2016-07-21 [1] CRAN (R 4.3.0)
AnnotationDbi * 1.64.1 2023-11-02 [1] Bioconductor
backports 1.4.1 2021-12-13 [1] CRAN (R 4.3.0)
beeswarm 0.4.0 2021-06-01 [1] CRAN (R 4.3.0)
Biobase * 2.62.0 2023-10-26 [1] Bioconductor
BiocGenerics * 0.48.1 2023-11-02 [1] Bioconductor
BiocManager 1.30.22 2023-08-08 [1] CRAN (R 4.3.0)
BiocStyle * 2.30.0 2023-10-26 [1] Bioconductor
Biostrings 2.70.2 2024-01-30 [1] Bioconductor 3.18 (R 4.3.2)
bit 4.0.5 2022-11-15 [1] CRAN (R 4.3.0)
bit64 4.0.5 2020-08-30 [1] CRAN (R 4.3.0)
bitops 1.0-7 2021-04-24 [1] CRAN (R 4.3.0)
blob 1.2.4 2023-03-17 [1] CRAN (R 4.3.0)
bslib 0.6.1 2023-11-28 [1] CRAN (R 4.3.1)
cachem 1.0.8 2023-05-01 [1] CRAN (R 4.3.0)
callr 3.7.5 2024-02-19 [1] CRAN (R 4.3.1)
cellranger 1.1.0 2016-07-27 [1] CRAN (R 4.3.0)
checkmate 2.3.1 2023-12-04 [1] CRAN (R 4.3.1)
cli 3.6.2 2023-12-11 [1] CRAN (R 4.3.1)
cluster 2.1.6 2023-12-01 [1] CRAN (R 4.3.1)
clustree * 0.5.1 2023-11-05 [1] CRAN (R 4.3.1)
codetools 0.2-19 2023-02-01 [1] CRAN (R 4.3.2)
colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.3.0)
cowplot 1.1.3 2024-01-22 [1] CRAN (R 4.3.1)
crayon 1.5.2 2022-09-29 [1] CRAN (R 4.3.0)
data.table * 1.15.0 2024-01-30 [1] CRAN (R 4.3.1)
DBI 1.2.2 2024-02-16 [1] CRAN (R 4.3.1)
DelayedArray 0.28.0 2023-11-06 [1] Bioconductor
deldir 2.0-2 2023-11-23 [1] CRAN (R 4.3.1)
digest 0.6.34 2024-01-11 [1] CRAN (R 4.3.1)
dotCall64 1.1-1 2023-11-28 [1] CRAN (R 4.3.1)
dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.3.1)
edgeR * 4.0.16 2024-02-20 [1] Bioconductor 3.18 (R 4.3.2)
ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.3.0)
evaluate 0.23 2023-11-01 [1] CRAN (R 4.3.1)
fansi 1.0.6 2023-12-08 [1] CRAN (R 4.3.1)
farver 2.1.1 2022-07-06 [1] CRAN (R 4.3.0)
fastDummies 1.7.3 2023-07-06 [1] CRAN (R 4.3.0)
fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.3.0)
fitdistrplus 1.1-11 2023-04-25 [1] CRAN (R 4.3.0)
forcats * 1.0.0 2023-01-29 [1] CRAN (R 4.3.0)
fs 1.6.3 2023-07-20 [1] CRAN (R 4.3.0)
future 1.33.1 2023-12-22 [1] CRAN (R 4.3.1)
future.apply 1.11.1 2023-12-21 [1] CRAN (R 4.3.1)
generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
GenomeInfoDb 1.38.6 2024-02-10 [1] Bioconductor 3.18 (R 4.3.2)
GenomeInfoDbData 1.2.11 2024-02-27 [1] Bioconductor
GenomicRanges 1.54.1 2023-10-30 [1] Bioconductor
getPass 0.2-4 2023-12-10 [1] CRAN (R 4.3.1)
ggbeeswarm 0.7.2 2023-04-29 [1] CRAN (R 4.3.0)
ggforce 0.4.2 2024-02-19 [1] CRAN (R 4.3.1)
ggplot2 * 3.5.0 2024-02-23 [1] CRAN (R 4.3.1)
ggraph * 2.1.0 2022-10-09 [1] CRAN (R 4.3.0)
ggrastr 1.0.2 2023-06-01 [1] CRAN (R 4.3.0)
ggrepel 0.9.5 2024-01-10 [1] CRAN (R 4.3.1)
ggridges 0.5.6 2024-01-23 [1] CRAN (R 4.3.1)
git2r 0.33.0 2023-11-26 [1] CRAN (R 4.3.1)
globals 0.16.2 2022-11-21 [1] CRAN (R 4.3.0)
glue * 1.7.0 2024-01-09 [1] CRAN (R 4.3.1)
goftest 1.2-3 2021-10-07 [1] CRAN (R 4.3.0)
graphlayouts 1.1.0 2024-01-19 [1] CRAN (R 4.3.1)
gridExtra 2.3 2017-09-09 [1] CRAN (R 4.3.0)
gtable 0.3.4 2023-08-21 [1] CRAN (R 4.3.0)
here * 1.0.1 2020-12-13 [1] CRAN (R 4.3.0)
highr 0.10 2022-12-22 [1] CRAN (R 4.3.0)
hms 1.1.3 2023-03-21 [1] CRAN (R 4.3.0)
htmltools 0.5.7 2023-11-03 [1] CRAN (R 4.3.1)
htmlwidgets 1.6.4 2023-12-06 [1] CRAN (R 4.3.1)
httpuv 1.6.14 2024-01-26 [1] CRAN (R 4.3.1)
httr 1.4.7 2023-08-15 [1] CRAN (R 4.3.0)
ica 1.0-3 2022-07-08 [1] CRAN (R 4.3.0)
igraph 2.0.2 2024-02-17 [1] CRAN (R 4.3.1)
IRanges * 2.36.0 2023-10-26 [1] Bioconductor
irlba 2.3.5.1 2022-10-03 [1] CRAN (R 4.3.2)
jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.3.0)
jsonlite 1.8.8 2023-12-04 [1] CRAN (R 4.3.1)
kableExtra * 1.4.0 2024-01-24 [1] CRAN (R 4.3.1)
KEGGREST 1.42.0 2023-10-26 [1] Bioconductor
KernSmooth 2.23-22 2023-07-10 [1] CRAN (R 4.3.2)
knitr 1.45 2023-10-30 [1] CRAN (R 4.3.1)
labeling 0.4.3 2023-08-29 [1] CRAN (R 4.3.0)
later 1.3.2 2023-12-06 [1] CRAN (R 4.3.1)
lattice 0.22-5 2023-10-24 [1] CRAN (R 4.3.1)
lazyeval 0.2.2 2019-03-15 [1] CRAN (R 4.3.0)
leiden 0.4.3.1 2023-11-17 [1] CRAN (R 4.3.1)
lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.3.1)
limma * 3.58.1 2023-11-02 [1] Bioconductor
listenv 0.9.1 2024-01-29 [1] CRAN (R 4.3.1)
lmtest 0.9-40 2022-03-21 [1] CRAN (R 4.3.0)
locfit 1.5-9.8 2023-06-11 [1] CRAN (R 4.3.0)
lubridate * 1.9.3 2023-09-27 [1] CRAN (R 4.3.1)
magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.0)
MASS 7.3-60.0.1 2024-01-13 [1] CRAN (R 4.3.1)
Matrix 1.6-5 2024-01-11 [1] CRAN (R 4.3.1)
MatrixGenerics 1.14.0 2023-10-26 [1] Bioconductor
matrixStats 1.2.0 2023-12-11 [1] CRAN (R 4.3.1)
memoise 2.0.1 2021-11-26 [1] CRAN (R 4.3.0)
mime 0.12 2021-09-28 [1] CRAN (R 4.3.0)
miniUI 0.1.1.1 2018-05-18 [1] CRAN (R 4.3.0)
munsell 0.5.0 2018-06-12 [1] CRAN (R 4.3.0)
nlme 3.1-164 2023-11-27 [1] CRAN (R 4.3.1)
org.Hs.eg.db * 3.18.0 2024-02-27 [1] Bioconductor
paletteer 1.6.0 2024-01-21 [1] CRAN (R 4.3.1)
parallelly 1.37.0 2024-02-14 [1] CRAN (R 4.3.1)
patchwork * 1.2.0 2024-01-08 [1] CRAN (R 4.3.1)
pbapply 1.7-2 2023-06-27 [1] CRAN (R 4.3.0)
pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.0)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.0)
plotly 4.10.4 2024-01-13 [1] CRAN (R 4.3.1)
plyr 1.8.9 2023-10-02 [1] CRAN (R 4.3.1)
png 0.1-8 2022-11-29 [1] CRAN (R 4.3.0)
polyclip 1.10-6 2023-09-27 [1] CRAN (R 4.3.1)
presto 1.0.0 2024-02-27 [1] Github (immunogenomics/presto@31dc97f)
prismatic 1.1.1 2022-08-15 [1] CRAN (R 4.3.0)
processx 3.8.3 2023-12-10 [1] CRAN (R 4.3.1)
progressr 0.14.0 2023-08-10 [1] CRAN (R 4.3.0)
promises 1.2.1 2023-08-10 [1] CRAN (R 4.3.0)
ps 1.7.6 2024-01-18 [1] CRAN (R 4.3.1)
purrr * 1.0.2 2023-08-10 [1] CRAN (R 4.3.0)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.0)
RANN 2.6.1 2019-01-08 [1] CRAN (R 4.3.0)
RColorBrewer * 1.1-3 2022-04-03 [1] CRAN (R 4.3.0)
Rcpp 1.0.12 2024-01-09 [1] CRAN (R 4.3.1)
RcppAnnoy 0.0.22 2024-01-23 [1] CRAN (R 4.3.1)
RcppHNSW 0.6.0 2024-02-04 [1] CRAN (R 4.3.1)
RCurl 1.98-1.14 2024-01-09 [1] CRAN (R 4.3.1)
readr * 2.1.5 2024-01-10 [1] CRAN (R 4.3.1)
readxl * 1.4.3 2023-07-06 [1] CRAN (R 4.3.0)
rematch2 2.1.2 2020-05-01 [1] CRAN (R 4.3.0)
reshape2 1.4.4 2020-04-09 [1] CRAN (R 4.3.0)
reticulate 1.35.0 2024-01-31 [1] CRAN (R 4.3.1)
rlang 1.1.3 2024-01-10 [1] CRAN (R 4.3.1)
rmarkdown 2.25 2023-09-18 [1] CRAN (R 4.3.1)
ROCR 1.0-11 2020-05-02 [1] CRAN (R 4.3.0)
rprojroot 2.0.4 2023-11-05 [1] CRAN (R 4.3.1)
RSpectra 0.16-1 2022-04-24 [1] CRAN (R 4.3.0)
RSQLite 2.3.5 2024-01-21 [1] CRAN (R 4.3.1)
rstudioapi 0.15.0 2023-07-07 [1] CRAN (R 4.3.0)
Rtsne 0.17 2023-12-07 [1] CRAN (R 4.3.1)
S4Arrays 1.2.0 2023-10-26 [1] Bioconductor
S4Vectors * 0.40.2 2023-11-25 [1] Bioconductor 3.18 (R 4.3.2)
sass 0.4.8 2023-12-06 [1] CRAN (R 4.3.1)
scales 1.3.0 2023-11-28 [1] CRAN (R 4.3.1)
scattermore 1.2 2023-06-12 [1] CRAN (R 4.3.0)
sctransform 0.4.1 2023-10-19 [1] CRAN (R 4.3.1)
sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
Seurat * 5.0.1.9009 2024-02-28 [1] Github (satijalab/seurat@6a3ef5e)
SeuratObject * 5.0.1 2023-11-17 [1] CRAN (R 4.3.1)
shiny 1.8.0 2023-11-17 [1] CRAN (R 4.3.1)
SingleCellExperiment 1.24.0 2023-11-06 [1] Bioconductor
sp * 2.1-3 2024-01-30 [1] CRAN (R 4.3.1)
spam 2.10-0 2023-10-23 [1] CRAN (R 4.3.1)
SparseArray 1.2.4 2024-02-10 [1] Bioconductor 3.18 (R 4.3.2)
spatstat.data 3.0-4 2024-01-15 [1] CRAN (R 4.3.1)
spatstat.explore 3.2-6 2024-02-01 [1] CRAN (R 4.3.1)
spatstat.geom 3.2-8 2024-01-26 [1] CRAN (R 4.3.1)
spatstat.random 3.2-2 2023-11-29 [1] CRAN (R 4.3.1)
spatstat.sparse 3.0-3 2023-10-24 [1] CRAN (R 4.3.1)
spatstat.utils 3.0-4 2023-10-24 [1] CRAN (R 4.3.1)
speckle * 1.2.0 2023-10-26 [1] Bioconductor
statmod 1.5.0 2023-01-06 [1] CRAN (R 4.3.0)
stringi 1.8.3 2023-12-11 [1] CRAN (R 4.3.1)
stringr * 1.5.1 2023-11-14 [1] CRAN (R 4.3.1)
SummarizedExperiment 1.32.0 2023-11-06 [1] Bioconductor
survival 3.5-8 2024-02-14 [1] CRAN (R 4.3.1)
svglite 2.1.3 2023-12-08 [1] CRAN (R 4.3.1)
systemfonts 1.0.5 2023-10-09 [1] CRAN (R 4.3.1)
tensor 1.5 2012-05-05 [1] CRAN (R 4.3.0)
tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.3.0)
tidygraph 1.3.1 2024-01-30 [1] CRAN (R 4.3.1)
tidyr * 1.3.1 2024-01-24 [1] CRAN (R 4.3.1)
tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.3.0)
tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.3.0)
timechange 0.3.0 2024-01-18 [1] CRAN (R 4.3.1)
tweenr 2.0.3 2024-02-26 [1] CRAN (R 4.3.1)
tzdb 0.4.0 2023-05-12 [1] CRAN (R 4.3.0)
utf8 1.2.4 2023-10-22 [1] CRAN (R 4.3.1)
uwot 0.1.16 2023-06-29 [1] CRAN (R 4.3.0)
vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.3.1)
vipor 0.4.7 2023-12-18 [1] CRAN (R 4.3.1)
viridis 0.6.5 2024-01-29 [1] CRAN (R 4.3.1)
viridisLite 0.4.2 2023-05-02 [1] CRAN (R 4.3.0)
whisker 0.4.1 2022-12-05 [1] CRAN (R 4.3.0)
withr 3.0.0 2024-01-16 [1] CRAN (R 4.3.1)
workflowr * 1.7.1 2023-08-23 [1] CRAN (R 4.3.0)
xfun 0.42 2024-02-08 [1] CRAN (R 4.3.1)
xml2 1.3.6 2023-12-04 [1] CRAN (R 4.3.1)
xtable 1.8-4 2019-04-21 [1] CRAN (R 4.3.0)
XVector 0.42.0 2023-10-26 [1] Bioconductor
yaml 2.3.8 2023-12-11 [1] CRAN (R 4.3.1)
zlibbioc 1.48.0 2023-10-26 [1] Bioconductor
zoo 1.8-12 2023-04-13 [1] CRAN (R 4.3.0)
[1] /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/library
──────────────────────────────────────────────────────────────────────────────
sessionInfo()R version 4.3.2 (2023-10-31)
Platform: aarch64-apple-darwin20 (64-bit)
Running under: macOS Sonoma 14.6.1
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: Australia/Melbourne
tzcode source: internal
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] readxl_1.4.3 org.Hs.eg.db_3.18.0 AnnotationDbi_1.64.1
[4] IRanges_2.36.0 S4Vectors_0.40.2 Biobase_2.62.0
[7] BiocGenerics_0.48.1 speckle_1.2.0 edgeR_4.0.16
[10] limma_3.58.1 patchwork_1.2.0 data.table_1.15.0
[13] RColorBrewer_1.1-3 kableExtra_1.4.0 clustree_0.5.1
[16] ggraph_2.1.0 Seurat_5.0.1.9009 SeuratObject_5.0.1
[19] sp_2.1-3 glue_1.7.0 here_1.0.1
[22] lubridate_1.9.3 forcats_1.0.0 stringr_1.5.1
[25] dplyr_1.1.4 purrr_1.0.2 readr_2.1.5
[28] tidyr_1.3.1 tibble_3.2.1 ggplot2_3.5.0
[31] tidyverse_2.0.0 BiocStyle_2.30.0 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] fs_1.6.3 matrixStats_1.2.0
[3] spatstat.sparse_3.0-3 bitops_1.0-7
[5] httr_1.4.7 tools_4.3.2
[7] sctransform_0.4.1 backports_1.4.1
[9] utf8_1.2.4 R6_2.5.1
[11] lazyeval_0.2.2 uwot_0.1.16
[13] withr_3.0.0 gridExtra_2.3
[15] progressr_0.14.0 cli_3.6.2
[17] spatstat.explore_3.2-6 fastDummies_1.7.3
[19] prismatic_1.1.1 labeling_0.4.3
[21] sass_0.4.8 spatstat.data_3.0-4
[23] ggridges_0.5.6 pbapply_1.7-2
[25] systemfonts_1.0.5 svglite_2.1.3
[27] sessioninfo_1.2.2 parallelly_1.37.0
[29] rstudioapi_0.15.0 RSQLite_2.3.5
[31] generics_0.1.3 ica_1.0-3
[33] spatstat.random_3.2-2 Matrix_1.6-5
[35] ggbeeswarm_0.7.2 fansi_1.0.6
[37] abind_1.4-5 lifecycle_1.0.4
[39] whisker_0.4.1 yaml_2.3.8
[41] SummarizedExperiment_1.32.0 SparseArray_1.2.4
[43] Rtsne_0.17 paletteer_1.6.0
[45] grid_4.3.2 blob_1.2.4
[47] promises_1.2.1 crayon_1.5.2
[49] miniUI_0.1.1.1 lattice_0.22-5
[51] cowplot_1.1.3 KEGGREST_1.42.0
[53] pillar_1.9.0 knitr_1.45
[55] GenomicRanges_1.54.1 future.apply_1.11.1
[57] codetools_0.2-19 leiden_0.4.3.1
[59] getPass_0.2-4 vctrs_0.6.5
[61] png_0.1-8 spam_2.10-0
[63] cellranger_1.1.0 gtable_0.3.4
[65] rematch2_2.1.2 cachem_1.0.8
[67] xfun_0.42 S4Arrays_1.2.0
[69] mime_0.12 tidygraph_1.3.1
[71] survival_3.5-8 SingleCellExperiment_1.24.0
[73] statmod_1.5.0 ellipsis_0.3.2
[75] fitdistrplus_1.1-11 ROCR_1.0-11
[77] nlme_3.1-164 bit64_4.0.5
[79] RcppAnnoy_0.0.22 GenomeInfoDb_1.38.6
[81] rprojroot_2.0.4 bslib_0.6.1
[83] irlba_2.3.5.1 vipor_0.4.7
[85] KernSmooth_2.23-22 colorspace_2.1-0
[87] DBI_1.2.2 ggrastr_1.0.2
[89] tidyselect_1.2.0 processx_3.8.3
[91] bit_4.0.5 compiler_4.3.2
[93] git2r_0.33.0 xml2_1.3.6
[95] DelayedArray_0.28.0 plotly_4.10.4
[97] checkmate_2.3.1 scales_1.3.0
[99] lmtest_0.9-40 callr_3.7.5
[101] digest_0.6.34 goftest_1.2-3
[103] spatstat.utils_3.0-4 presto_1.0.0
[105] rmarkdown_2.25 XVector_0.42.0
[107] htmltools_0.5.7 pkgconfig_2.0.3
[109] MatrixGenerics_1.14.0 highr_0.10
[111] fastmap_1.1.1 rlang_1.1.3
[113] htmlwidgets_1.6.4 shiny_1.8.0
[115] farver_2.1.1 jquerylib_0.1.4
[117] zoo_1.8-12 jsonlite_1.8.8
[119] RCurl_1.98-1.14 magrittr_2.0.3
[121] GenomeInfoDbData_1.2.11 dotCall64_1.1-1
[123] munsell_0.5.0 Rcpp_1.0.12
[125] viridis_0.6.5 reticulate_1.35.0
[127] stringi_1.8.3 zlibbioc_1.48.0
[129] MASS_7.3-60.0.1 plyr_1.8.9
[131] parallel_4.3.2 listenv_0.9.1
[133] ggrepel_0.9.5 deldir_2.0-2
[135] Biostrings_2.70.2 graphlayouts_1.1.0
[137] splines_4.3.2 tensor_1.5
[139] hms_1.1.3 locfit_1.5-9.8
[141] ps_1.7.6 igraph_2.0.2
[143] spatstat.geom_3.2-8 RcppHNSW_0.6.0
[145] reshape2_1.4.4 evaluate_0.23
[147] BiocManager_1.30.22 tzdb_0.4.0
[149] tweenr_2.0.3 httpuv_1.6.14
[151] RANN_2.6.1 polyclip_1.10-6
[153] future_1.33.1 scattermore_1.2
[155] ggforce_0.4.2 xtable_1.8-4
[157] RSpectra_0.16-1 later_1.3.2
[159] viridisLite_0.4.2 beeswarm_0.4.0
[161] memoise_2.0.1 cluster_2.1.6
[163] timechange_0.3.0 globals_0.16.2